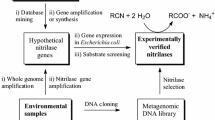
Abstract
Escherichia coli strains expressing different nitrilases transformed nitriles or KCN. Six nitrilases (from Aspergillus niger (2), A. oryzae, Neurospora crassa, Arthroderma benhamiae, and Nectria haematococca) were arylacetonitrilases, two enzymes (from A. niger and Penicillium chrysogenum) were cyanide hydratases and the others (from P. chrysogenum, P. marneffei, Gibberella moniliformis, Meyerozyma guilliermondi, Rhodococcus rhodochrous, and R. ruber) preferred (hetero)aromatic nitriles as substrates. Promising nitrilases for the transformation of industrially important substrates were found: the nitrilase from R. ruber for 3-cyanopyridine, 4-cyanopyridine and bromoxynil, the nitrilases from N. crassa and A. niger for (R,S)-mandelonitrile, and the cyanide hydratase from A. niger for KCN and 2-cyanopyridine.

Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Abbreviations
- aa:
-
Amino acid
- NitAb:
-
Nitrilase from Arthroderma benhamiae CBS 112371 (GenBank: EFE30690)
- NitAn:
-
Nitrilase from Aspergillus niger CBS 513.88 (GenBank: CAK46742)
- NitAn1:
-
Nitrilase from Aspergillus niger K10 (CCF 3411 GenBank: ABX75546)
- NitAn2:
-
Nitrilase from Aspergillus niger CBS 513.88 (GenBank: CAK47246)
- NitAo:
-
Nitrilase from Aspergillus oryzae RIB40 (GenBank: BAE63579)
- NitGm:
-
Nitrilase from Giberella moniliformis (GenBank: ABF83489)
- NitMg:
-
Nitrilase from Meyerozyma guilliermondi (Pichia guilliermondi) ATCC 6260 (NCBI Reference Sequence: XP_001482890)
- NitNc:
-
Nitrilase from Neurospora crassa OR74A (GenBank: CAD70472)
- NitNh:
-
Nitrilase from Nectria haematococca mpVI 77-13-4 (GenBank: EEU45207)
- NitPc1:
-
Nitrilase from Penicillium chrysogenum Wisconsin 54-1255 (NCBI Reference Sequence: XP_002562104)
- NitPc2:
-
Nitrilase from Penicillium chrysogenum Wisconsin 54-1255 (NCBI Reference Sequence: XP_002565836)
- NitPm:
-
Nitrilase from Penicillium marneffei ATCC18224 (NCBI Reference Sequence: XP_002144951)
- NitRr1:
-
T365S/L366I variant of nitrilase from Rhodococcus ruber PA-34 (EMBL Bank: HF543938)
- NitRr2:
-
T365S/L366I variant of nitrilase from Rhodococcus ruber NHB-2 (EMBL Bank: HF543939)
- NitRr3:
-
T365S/L366I variant of nitrilase from Rhodococcus rhodochrous NDB 1165 (EMBL Bank: HF543940)
References
Thimann, K. V., & Mahadevan, S. (1964). Nitrilase I. Occurrence, preparation and general properties of enzyme. Archives of Biochemistry and Biophysics, 105, 133–141.
O’Reilly, C., & Turner, P. D. (2003). The nitrilase family of CN hydrolysing enzymes—a comparative study. Journal of Applied Microbiology, 95, 1161–1174.
Thuku, R. N., Brady, D., Benedik, M. J., & Sewell, B. T. (2009). Microbial nitrilases: versatile, spiral forming, industrial enzymes. Journal of Applied Microbiology, 106, 703–727.
Martínková, L., & Křen, V. (2010). Biotransformations with nitrilases. Current Opinion in Chemical Biology, 14, 130–137.
Kobayashi, M., & Shimizu, S. (1994). Versatile nitrilases: Nitrile-hydrolysing enzymes. FEMS Microbiology Letters, 120, 217–223.
Raczynska, J. E., Vorgias, C. E., Antranikian, G., & Rypniewski, W. (2011). Crystallographic analysis of a thermoactive nitrilase. Journal of Structural Biology, 173, 294–302.
Yeom, S.-J., Kim, H.-J., Lee, J.-K., Kim, D.-E., & Oh, D.-K. (2008). An amino acid at position 142 in nitrilase from Rhodococcus rhodochrous ATCC 33278 determines the substrate specificity for aliphatic and aromatic nitriles. Biochemical Journal, 415, 401–407.
Seffernick, J. L., Samanta, S. K., Louie, T. M., Wackett, L. P., & Subramanian, M. (2009). Investigative mining of sequence data for novel enzymes: A case study with nitrilases. Journal of Biotechnology, 143, 17–26.
Robertson, D. E., Chaplin, J. A., DeSantis, G., Podar, M., Madden, M., Chi, E., et al. (2004). Exploring nitrilase sequence space for enantioselective catalysis. Applied and Environmental Microbiology, 70, 2429–2436.
Kaplan, O., Bezouška, K., Malandra, A., Veselá, A. B., Petříčková, A., Felsberg, J., et al. (2011). Genome mining for the discovery of new nitrilases in filamentous fungi. Biotechnology Letters, 33, 309–312.
Petříčková, A., Veselá, A. B., Kaplan, O., Kubáč, D., Uhnáková, B., Malandra, A., et al. (2012). Purification and characterization of heterologously expressed nitrilases from filamentous fungi. Applied Microbiology and Biotechnology, 93, 1553–1561.
Veselá, A. B., Petříčková, A., Weyrauch, P., & Martínková, L. (2012). Heterologous expression, purification and characterization of arylacetonitrilases from Nectria haematococca and Arthroderma benhamiae. Biocatalysis and Biotransformation,. doi:10.3109/10242422.2012.758117.
Komeda, H., Hori, Y., Kobayashi, M., & Shimizu, S. (1996). Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proceedings of the National Academy of Sciences of the United States of America, 93, 10572–10577.
Bhalla, T. C., Miura, A., Wakamoto, A., Ohba, Y., & Furuhashi, K. (1992). Asymmetric hydrolysis of alpha aminonitriles to optically active amino acids by a nitrilase of Rhodococcus rhodochrous PA-34. Applied Microbiology and Biotechnology, 37, 184–190.
Bhalla, T. C., & Kumar, H. (2005). Nocardia globerula NHB-2: A versatile nitrile-degrading organism. Canadian Journal of Microbiology, 51, 705–708.
Prasad, S., Misra, A., Jangir, V. P., Awasthi, A., Raj, J., & Bhalla, T. C. (2007). A propionitrile-induced nitrilase of Rhodococcus sp. NDB 1165 and its application in nicotinic acid synthesis. World Journal of Microbiology and Biotechnology, 23, 345–353.
Veselá, A. B., Pelantová, H., Šulc, M., Macková, M., Lovecká, P., Thimová, M., et al. (2012). Biotransformation of benzonitrile herbicides via the nitrile hydratase-amidase pathway in rhodococci. Journal of Industrial Microbiology and Biotechnology, 39, 1811–1819.
Bhalla, T. C., Aoshima, M., Misawa, S., Muramatsu, R., & Furuhashi, K. (1995). The molecular cloning and sequencing of the nitrilase gene of Rhodococcus rhodochrous PA-34. Acta Biotechnologica, 15, 297–306.
Nagasawa, T., Mauger, J., & Yamada, H. (1990). A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Purification and characterization. European Journal of Biochemistry, 194, 765–772.
Yamamoto, K., Fujimatsu, I., & Komatsu, K.-I. (1992). Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. Journal of Fermentation and Bioengineering, 73, 425–430.
Zhang, Z.-J., Xu, J.-H., He, Y.-C., Ouyang, L.-M., & Liu, Y.-Y. (2011). Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(-)-mandelic acid production. Bioprocess and Biosystems Engineering, 34, 315–322.
Banerjee, A., Dubey, S., Kaul, P., Barse, B., Piotrowski, M., & Banerjee, U. C. (2009). Enantioselective nitrilase from Pseudomonas putida. Cloning, heterologous expression, and bioreactor studies. Molecular Biotechnology, 41, 35–41.
Kiziak, C., Conradt, D., Stolz, A., Mattes, R., & Klein, J. (2005). Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology, 151, 3639–3648.
Chmura, A., Shapovalova, A. A., van Pelt, S., van Rantwijk, F., Tourova, T. P., Muyzer, G., et al. (2008). Utilization of arylaliphatic nitriles by haloalkaliphilic Halomonas nitrilicus sp. nov. isolated from soda soils. Applied Microbiology and Biotechnology, 81, 371–378.
Zhu, D., Mukherjee, C., Yang, Y., Rios, B. E., Gallagher, D. T., Smith, N. N., et al. (2008). A new nitrilase from Bradyrhizobium japonicum USDA 110. Gene cloning, biochemical characterization and substrate specificity. Journal of Biotechnology, 133, 327–333.
Kiziak, C., Klein, J., & Stolz, A. (2007). Influence of different carboxy-terminal mutations on the substrate-, reaction- and enantiospecificity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Protein Engineering, Design & Selection, 20, 385–396.
Kiziak, C., & Stolz, A. (2009). Identification of amino acid residues responsible for the enantioselectivity and amide formation capacity of the arylacetonitrilase from Pseudomonas fluorescens EBC191. Applied and Environmental Microbiology, 75, 5592–5599.
Thuku, R. N., Weber, B. W., Varsani, A., & Sewell, B. T. (2007). Post-translational cleavage of recombinantly expressed nitrilase from Rhodococcus rhodochrous J1 yields a stable, active helical form. FEBS Journal, 274, 2099–2108.
Luo, H., Fan, L., Chang, Y. H., Ma, J. W., Yu, H. M., & Shen, Z. Y. (2010). Gene cloning, overexpression, and characterization of the nitrilase from Rhodococcus rhodochrous tg1-A6 in E. coli. Applied Biochemistry and Biotechnology, 160, 393–400.
Goldlust, A., & Bohak, Z. (1989). Induction, purification, and characterization of the nitrilase of Fusarium oxysporum f. sp. melonis. Biotechnology and Applied Biochemistry, 11, 581–601.
Basile, L. J., Willson, R. C., Sewell, B. T., & Benedik, M. J. (2008). Genome mining of cyanide-degrading nitrilases from filamentous fungi. Applied Microbiology and Biotechnology, 80, 427–435.
Almatawah, Q. A., Cramp, R., & Cowan, D. A. (1999). Characterization of an inducible nitrilase from a thermophilic bacillus. Extremophiles, 3, 283–291.
Acknowledgments
This study was supported via projects P504/11/0394 (Czech Science Foundation), TA01021368 (Technology Agency of the Czech Republic) and internal project RVO61388971 (Institute of Microbiology).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2013_9656_MOESM1_ESM.doc
Supplemental file 1 Nucleotide sequences of the nitrilase genes adapted for expression in Escherichia coli using the Generay (Shanghai, China) software (DOC 30 kb)
Rights and permissions
About this article
Cite this article
Kaplan, O., Veselá, A.B., Petříčková, A. et al. A Comparative Study of Nitrilases Identified by Genome Mining. Mol Biotechnol 54, 996–1003 (2013). https://doi.org/10.1007/s12033-013-9656-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-013-9656-6