Abstract
Small ubiquitin-related modifier (SUMO) fusion system has been shown to be efficient for enhancing expression and preventing degradation of the target protein. We showed herein that SUMO fusion to human keratinocyte growth factor 2 (hKGF-2) gene was feasible and it significantly enhanced protein expression and its efficiency. The fusion DNA fragment composed of SUMO gene, which was fused to hexahistidine tag, and hKGF-2 gene was amplified by PCR and inserted into the expression vector pET28a to construct the recombinant plasmid, pET28a-SUMO-hKGF-2. The plasmid was then transformed into Escherichia coli RosettaTM2(DE3), and the recombinant fusion protein SUMO-hKGF-2 was expressed at 30°C for 6 h, with the induction of IPTG at the final concentration of 0.4 mM. The expression level of the fusion protein was up to 30% of the total cellular protein. The fusion protein was purified by Ni-NTA affinity chromatography. After desalting by Sephadex G-25 size exclusion chromatography, the hexahistidine-SUMO-hKGF-2 was digested by SUMO proteases. The recombinant hKGF-2 was purified again with Ni-NTA column and the purity was about 95% with a total yield of 13.9 mg/l culture. The result of mitogenicity assay suggests that the recombinant hKGF-2 can significantly promote the proliferation of normal rat kidney epithelial (NRK-52E) cells.
Similar content being viewed by others
Introduction
Keratinocyte growth factor 2 (KGF-2), also known as fibroblast growth factor 10 (FGF-10), is a member of the fibroblast growth factor family, which includes a minimum of 23 related peptides. The protein sequence and biological function of KGF-2 are similar to that of KGF-1, or fibroblast growth factor 7 (FGF-7) [1]. KGF-2 is produced by fibroblasts of the dermis, the granulation tissue, and the intraepithelial γδT cells; it acts specifically on cells of epithelial origin, including skin keratinocytes, and intestinal epithelial cells, by binding to FGF receptor 2-IIIb (FGFR2-IIIb) [2, 3]. The accelerating effect of KGF-2 on the rate of re-epithelialization in various animal models has been demonstrated [4–6]. Furthermore, KGF-2’s protective properties against radiation and oxidative stress have also been demonstrated in recent studies [7, 8]. These preliminary data undeniably suggests KGF-2 as a promising candidate for the treatment of epithelial damage. Currently, KGF-2 is already being used in clinical trials for the treatment of venous ulcers [9] and oral mucositis, a severe oral ulceration induced by chemotherapy and/or radiotherapy. Large scale production of this promising therapeutic protein is demanded.
Nevertheless, high quality therapeutic proteins require rapid and efficient strategies for protein expression and purification. Currently, fusion protein technology is an effective way to achieve the goal by enhancing expression, decreasing the proteolytic degradation, improving solubility, and facilitating purification of the recombinant proteins [10, 11]. Despite the current advancements in fusion protein technology, it is still burdened with flaws. Most of the fusion systems produce non-native N-terminal amino acids during the cleavage process of the fusion tag, which may lead to serious effects on the biological activities of many proteins. Small ubiquitin-related modifier (SUMO) family proteins are present in all eukaryotes but absent from prokaryotes. Sumoylation has crucial roles in regulation of gene expression, control of nucleocytoplasmic signaling, and faithful replication of a large and complex genome [12, 13]. SUMO fusion system could overcome the problems described above, and has been used to facilitate efficient expression and purification of several proteins in E. coli [14, 15]. The relative ease of maturation of SUMO fusions allows the target protein to be isolated using a standardized purification procedure; and the properties of the SUMO hydrolase, which recognizes the tertiary structure of the SUMO tag, allow the target protein to be produced with a completely native sequence. Butt et al. [16] have observed that 100 SUMO fusions have been isolated without cleavage within the partner protein. SUMO fusion system has also been successfully applied to some difficult-to-express proteins including matrix metalloprotease (MMP13), and SARS-CoV proteins [14, 15].
KGFs have been expressed in different systems. Luo et al. [17] have used GST-fusion technology and high MgCl2 concentration to improve expression level of KGF-1 in E. coli. Laird et al. [18] have observed that only the trunked form of KGF-2 was robustly expressed in a protease-deficient E. coli host. Recently, Wang et al. [19] demonstrated that recombinant human KGF-2 could be expressed in a yeast strain Pichia pastoris. However, the targeted protein with a molecular weight of 19.3 kDa had a significantly lower expression in the supernatant of transformants, induced in normal conditions, than a peptide with molecular weight of 17 kDa, which is speculated to be the degraded product of recombinant human KGF-2. This result suggests that the recombinant human KGF-2 may be susceptible to proteolytic degradation. Therefore, here we attempt to use a hexahistidine-SUMO fusion strategy for expression and purification of recombinant human KGF-2 in E. coli, aiming at decreasing the chance of proteolytic degradation, enhancing expression and simplifying purification of the target protein with Ni-NTA chromatography.
Experimental
Reagent
Pyrobest DNA Polymerase, restriction enzymes, polymerase chain reaction (PCR) purification kit, gel extraction kit, and plasmid miniprep kit were purchased from Takara (Dalian, China). Ni-NTA agarose and Sephadex G-25 were purchased from Invitrogen (Carlsbad, California, USA). Monoclonal mouse anti-human KGF-2 antibody was purchased from R&D systems (Minneapolis, MN, USA). Methylthiazoletetrazolium (MTT) was from Sigma–Aldrich (St. Louis, MO, USA).
SUMO protease was produced in our laboratory. Normal rat kidney epithelial (NRK-52E) cells was originated from American Type Culture Collection (ATCC, Rockville, MD, USA), and provided by Biopharmaceutical Research and Development Center of Jinan University.
Construction of SUMO-hKGF-2 Fusion Protein Expression Vector
Four primers were synthesized according to the DNA sequences of SUMO (GenBank accession number U27233) and hKGF-2 (GenBank accession number AB002097) (P1: CGTGGGATCCTCGGACTCAGAAGTCAATCA, P2: CAAGGGCTTGACCACCAATCTGTTCTCTGT, P3: GATTGGTGGTCAAGCCCTTGGTCAGGACAT, P4: CACGCTCGAGCTATGAGTGTACCACCATTG). Three-step polymerase chain reaction (PCR) was conducted to obtain fusion gene of SUMO-hKGF-2. First, we amplified SUMO gene fragment (F1) from a pET28a/SUMO-MT (constructed in our laboratory) plasmid, which contains hexahistidine tag for purification, using P1 and P2 as the forward primer and reverse primer. Second, hKGF-2 gene fragment (F2) was obtained from pET3c/hKGF-2 (constructed in our laboratory) plasmid using P3 and P4 as the primer pair. Finally, using F1 and F2 as the templates, the full length fusion gene was amplified using P1 and P4 as the forward primer and reverse primer. The product from the last round of PCR was cut with BamH I and Xho I (the restriction enzyme sites in the primers are indicated as italic letters), then it was ligated into previously digested vector pET28a to create the SUMO-hKGF-2 fusion protein expression vector, pET28a/SUMO-hKGF-2. Automated DNA sequencing was performed to confirm the accuracy of the inserted DNA segment.
Induction and Expression of SUMO-hKGF-2 Fusion Protein
The recombinant plasmid pET28a/SUMO-hKGF-2 harboring the accurate sequence of SUMO-hKGF-2 fusion gene was transferred into E. coli RosettaTM2(DE3) (Novagen). Each of the transformed colonies was grown in 4 ml Luria broth (LB) medium containing 50 μg/ml kanamycin and 34 μg/ml chloromycetin at 37°C. When the absorbance at 600 nm of the culture reached 0.6, the target protein was induced by adding isopropyl-b-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After induction at 37°C for 4 h, samples were prepared for expression analysis by SDS-PAGE. The colony with the highest expression level determined by SDS-PAGE was selected as seed strain in the subsequent scale-up culture.
The effects of IPTG concentration, temperature, and induction time on the expression level of target protein were studied to determine the optimal induction conditions. The culture was first induced at 0.2, 0.4, 0.6, 0.8, and 1 mM of IPTG concentration, respectively, at 37°C for 4 h to select the optimal IPTG concentrations. Then the induction was performed at optimal IPTG concentration at 37°C for 3, 4, 5, 6, 7, and 8 h, at 30°C for 4, 6, 8, 10, and 12 h, and at 20°C for 12, 14, 16, 18, 20, 22, and 24 h, respectively.
We further analyzed the expression of target protein at the selected optimal induction conditions. Briefly, cells were collected by centrifugation after induction and the cell pellets were resuspended in 20 mM PBS buffer supplemented with lysozyme. The suspensions were freeze–thawed for three times and centrifuged at 16,000 rpm (31,554g) for 20 min at 4°C. Samples obtained from the supernatant and pellets were loaded on SDS-PAGE gels for target protein expression analysis.
Purification of SUMO-hKGF-2 Fusion Protein
The cell extract was applied to a Ni-NTA agarose column equilibrated with 20 mM PBS buffer (pH 8.0). The resin was washed with 50 ml PBS (pH 8.0). The contaminated proteins were eluted in succession with PBS containing 30 mM imidazole (pH 8.0) and PBS containing 30 mM imidazole (pH 6.0). Bound proteins were eluted with 20 mM PBS containing 300 mM imidazole (pH 8.0). Fractions were pooled and further desalted with Sephadex G-25 column. The purity of SUMO-hKGF-2 fusion protein was assessed using SDS-PAGE, and the immunogenic activity of the fusion protein was detected by Western blotting.
Cleavage of SUMO-hKGF-2 Fusion Protein and Purification of hKGF-2
The purified fusion protein was mixed with SUMO protease at a molar ratio of 6,000:1. The mixture was incubated at 30°C for 1.5 h. The cleaved sample was applied to the Ni-NTA resin to separate the recombinant hKGF-2 from the resin-bound proteins including SUMO-hkGF-2, SUMO and SUMO protease. The immunogenic activity of hKGF-2 was also detected by western blotting. The concentration of hKGF-2 was evaluated with enhanced BCA protein assay Kit.
Bioassay of Mitogenic Activity of Recombinant hKGF-2
NRK-52E cells were grown in DMEM supplemented with 10% fetal bovine serum, 100 U/ml ampicillin and 100 U/ml streptomycin. When the culture reached the mid-logarithm time, cells were transferred to a 96-well plate and incubated at 37°C for 24 h. The medium was replaced with DMEM supplemented with 0.4% fetal bovine serum and the cells were cultured for 24 h. The cells were treated with recombinant hKGF-2 and SUMO-hKGF-2 diluted with serum free DMEM to a serial of concentrations ranged from 0.01 to 100 ng/ml, and incubated for 48 h. The number of viable cells was determined by adding 20 μl methylthiazoleterazolium (MTT) (5 mg/ml) to each well and incubated for 4 h. After removal of the medium, 150 μl dimethyl sulfoxide (DMSO) was added to each well. The plate was kept at room temperature for 30 min. The absorbance was measured at 570 nm immediately.
Results
Expression of sSUMO-hKGF-2 Fusion Protein
Three steps of PCR were performed to obtain the DNA fragment coding fusion protein composed of SUMO and hKGF-2 (Fig. 1). The product from the last round PCR was digested with BamH I and Xho I, and inserted in the expression vector pET-28a to create the recombinant plasmid pET28a/SUMO-hKGF-2, which was confirmed by automated DNA sequencing and transformed into E. coli RosettaTM2(DE3).
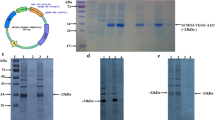
After induction with 1 mM IPTG at 37°C for 4 h, a protein band with molecular weight of approximately 38 kDa corresponding to the predicted size of SUMO-hKGF-2 fusion protein was obtained (Fig. 2 lane 4). The expression level of target protein induced at 0.4 mM of IPTG was comparable to that induced at 0.6, 0.8 and 1 mM, and higher than that induced at 0.2 mM (data not shown). Therefore, 0.4 mM of IPTG was selected for further experiments. Time- and temperature-dependent induction was also investigated. The expression level of the fusion protein reached maximum when induced at 37°C for 4 h, at 30°C for 6 h, and at 20°C for 12 h, respectively. Analysis of expression of target protein at the above conditions indicated that the soluble expression level of SUMO-hKGF-2 fusion protein was 26.9% at 37°C for 4 h, 31.3% at 30°C for 6 h, and 31.6% at 20°C for 12 h, respectively (Fig. 3). Based on these results, the optimal expression condition was selected as induction with 0.4 mM IPTG in culture medium at 30°C for 6 h.
Temperature effect on expression of SUMO-hKGF-2 fusion protein. M low molecular protein marker; 1, 4, 7 RossetaTM2(DE3)/pET28a/SUMO-hKGF-2 before induction; 2, 5, 8 supernatant of RossetaTM2(DE3)/pET28a/SUMO-hKGF-2 after induction; 3, 6, 9 sediment of RossetaTM2(DE3)/pET28a/SUMO-hKGF-2 after induction
Purification of SUMO-hKGF-2 Fusion Protein
After induction, the cell pellet was sonicated and the supernatant was applied to an affinity column. Proteins without His tag was first removed from the Ni-NTA resin by washing with PBS containing low concentration of imidazole, and the fusion protein with His tag was then eluted with PBS containing high concentration of imidazole. The eluted fraction was further desalted by applying to a Sephadex G-25 column. Detected by SDS-PAGE, the purity of SUMO-hKGF-2 after desalting was 94% (Fig. 4. Panel A, lane 1).
Panel A purification and digestion of SUMO-hKGF-2 (Coomassie blue staining). M low molecular protein marker; 1 SUMO-hKGF-2 fusion protein after Ni–NTA affinity chromatography and desalted by Sephadex G-25 chromatography; 2 SUMO-hKGF-2 fusion protein after Ni-NTA affinity chromatography. 3 SUMO-hKGF-2 before digestion; 4 SUMO-hKGF-2 protein after digestion at 30°C for 1.5 h; 5 purified hKGF-2. Panel B western blots of SUMO-hKGF-2 and hKGF-2. 1 purified SUMO-hKGF-2; 2 purified hKGF-2
Cleavage of SUMO-hKGF-2 and Purification of hKGF-2
The purified SUMO-hKGF-2 fusion protein was cleaved with SUMO protease to release hKGF-2. The result of SDS-PAGE showed that most of fusion protein was cleaved within 1.5 h at 30°C (Fig. 4. Panel A, lane 4). The reaction mixture was then reloaded onto the Ni-NTA resin for purification. The relieved hKGF-2 was obtained in the flow-through, whereas the His-containing uncleaved fusion protein and His-SUMO tag were retained by the resin. The purity of hKGF-2 determined by optic density scanning in SDS-PAGE gel was about 95% (Fig. 4. Panel A, lane 5). The summary of purification is presented in Table 1. The final yield of purified recombinant hKGF-2 was about 13 mg/l culture. Further characterization using western blot showed that both of the purified recombinant hKGF-2 and SUMO-hKGF-2 fusion protein could be recognized by an anti-human KGF-2 antibody (Fig. 4. Panel B).
Mitogenic Activity of Recombinant hKGF-2
The result of mitogenic assay showed that hKGF-2 cleaved from SUMO-hKGF-2 fusion protein could significantly stimulate the proliferation of NRK-52E cells. The result was similar with what was reported by Igarashi et al. [1], who assessed the mitogenic activity of recombinant hKGF-2 using mouse epidermal keratinocytes (Balb/MK) in the presence of 1 μg/ml heparin. Although NRK-52E cell proliferation was also stimulated by SUMO-hKGF-2, the mitogenic activity was markedly lower than that of hKGF-2, presumably due to the conformation change or shielding of the hKGF-2 active sites by SUMO (Fig. 5).
Discussion
hKGF-2 is one of the major mitogens and cytoprotectors of epithelial cells that has therapeutic potential in healing wound, curing ulcer and colitis. Increasing clinical interests have been shown in producing high quantity and quality recombinant hKGF-2. Expression of hKGF-2 in E. coli without tag obtained only 10–15% of aim product of total protein [20]. Jang [21] used GST-fusion to produce KGF-2, but without description of expression level. Other studies used various expression vectors and hosts obtained only 4–20% expression levels [22]. Low expression level due to susceptibility to proteolytic degradation and complicated purification procedure prevented the efficient production of functional hKGF-2 with high purity.
The lack of efficient methods for protein expression manifested the need to use a different approach. Recently, SUMO was found to have the abilities of enhancing protein expression level, improving protein folding, and protecting the protein from degradation by functioning as a chaperone and a nucleation site for protein folding. Some difficult-to-express proteins have been successfully expressed using SUMO as a novel fusion partner. The goal of this study is to express recombinant hKGF-2 with SUMO fusion in E. coli.
Different host strains including BL21(DE3), BL21(DE3)pLyS, and RosettaTM2(DE3) were used to express SUMO-hKGF-2 fusion protein. The corresponding expression level was approximately 12, 14, and 30% of the total cellular proteins, respectively. Accumulating evidence suggests that an excess of rare codons decrease expression level of heterologous proteins in E. coli. Moreover, sequence analysis of SUMO-hKGF-2 fusion gene showed that it contained up to 20% of codons rarely used in E. coli, suggesting that rare codons may restrict the expression of fusion protein. To overcome this problem, Rosetta™ 2(DE3) host strain which is a BL21 derivative was designed to enhance the expression of eukaryotic proteins that contain codons rarely used in E. coli by supplying tRNAs for seven rare codons (AGA, AGG, AUA, CUA, GGA, CCC, and CGG) on a compatible chloramphenicol-resistant plasmid. The results showed that Rosetta™ 2(DE3) strain is suitable for high-level expression of SUMO-hKGF-2 fusion protein.
We also optimized expression conditions, including IPTG concentration, temperature, and induction time, to reach high-level soluble expression of SUMO-hKGF-2 fusion protein. As a His tag was fused at N-terminal of SUMO-hKGF-2, one-step purification with Ni-NTA affinity chromatography was used to purify fusion protein. Moreover, since both SUMO fusion protein and SUMO contained an N-terminal His tag, hKGF-2 could be readily and rapidly purified from the cleaved SUMO-hKGF-2 samples by re-applying the cleaved samples to the Ni-NTA column. Another important property of hKGF-2 expressed by our SUMO fusion strategy is that it is biofunctional, indicating its feasibility of further large scale production.
In summary, our studies demonstrated that fusion of SUMO to the N-terminal of hKGF-2 enhanced expression. Furthermore, this method also protects hKGF-2 from proteolytic degradation in E. coli cells. Hexahistidine-tagged SUMO-fusions facilitated rapid and efficient purification of biological functional recombinant hKGF-2. These results manifest the essentiality of SUMO-fusion proteins by establishing a foundation for which large-scale production of hKGF-2 could be made possible. The practicality of this technique is very well worth considering for potential clinical practices.
References
Igarashi, M., Finch, P. W., & Aaronson, S. A. (1998). Characterization of recombinant human fibroblast growth factor (FGF)-10 reveals functional similarities with keratinocyte growth factor (FGF-7). The Journal of Biological Chemistry, 273, 13230–13235. doi:10.1074/jbc.273.21.13230.
Jameson, J., Ugarte, K., Chen, N., Yachi, P., Fuchs, E., Boismenu, R., et al. (2002). A role for skin gammadelta T cells in wound repair. Science, 296, 747–749. doi:10.1126/science.1069639.
Miki, T., Bottaro, D. P., Fleming, T. P., Smith, C. L., Burgess, W. H., Chan, A. M., et al. (1992). Determination of ligand-binding specificity by alternative splicing: Two distinct growth factor receptors encoded by a single gene. Proceedings of the National Academy of Sciences of the United States of America, 89, 246–250. doi:10.1073/pnas.89.1.246.
Xia, Y. P., Zhao, Y., Marcus, J., Jimenez, P. A., Ruben, S. M., Moore, P. A., et al. (1999). Effects of keratinocyte growth factor-2 (KGF-2) on wound healing in an ischaemia-impaired rabbit ear model and on scar formation. The Journal of Pathology, 188, 431–438. doi :10.1002/(SICI)1096-9896(199908)188:4<431::AID-PATH362>3.0.CO;2-B.
Jimenez, P. A., & Rampy, M. A. (1999). Keratinocyte growth factor-2 accelerates wound healing in incisional wounds. The Journal of Surgical Research, 81, 238–242. doi:10.1006/jsre.1998.5501.
Han, D. S., Li, F., Holt, L., Connolly, K., Hubert, M., Miceli, R., et al. (2000). Keratinocyte growth factor-2 (FGF-10) promotes healing of experimental small intestinal ulceration in rats. American Journal of Physiology. Gastrointestinal and Liver Physiology, 279, G1011–G1022.
Braun, S., auf dem Keller, U., Steiling, H., & Werner, S. (2004). Fibroblast growth factors in epithelial repair and cytoprotection. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 359, 753–757. doi:10.1098/rstb.2004.1464.
Upadhyay, D., Chang, W., Wei, K., Gao, M., & Rosen, G. D. (2007). Fibroblast growth factor-10 prevents H2O2-induced cell cycle arrest by regulation of G1 cyclins and cyclin dependent kinases. FEBS Letters, 581, 248–252. doi:10.1016/j.febslet.2006.12.020.
Robson, M. C., Phillips, T. J., Falanga, V., Odenheimer, D. J., Parish, L. C., Jensen, J. L., et al. (2001). Randomized trial of topically applied repifermin (recombinant human keratinocyte growth factor-2) to accelerate wound healing in venous ulcers. Wound Repair and Regeneration, 9, 347–352. doi:10.1046/j.1524-475x.2001.00347.x.
De Marco, V., Stier, G., Blandin, S., & de Marco, A. (2004). The solubility and stability of recombinant proteins are increased by their fusion to NusA. Biochemical and Biophysical Research Communications, 322, 766–771. doi:10.1016/j.bbrc.2004.07.189.
Pryor, K. D., & Leiting, B. (1997). High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expression and Purification, 10, 309–319. doi:10.1006/prep.1997.0759.
Seeler, J. S., & Dejean, A. (2003). Nuclear and unclear functions of SUMO. Nature Reviews. Molecular Cell Biology, 4, 690–699. doi:10.1038/nrm1200.
Malakhov, M. P., Mattern, M. R., Malakhova, O. A., Drinker, M., Weeks, S. D., & Butt, T. R. (2004). SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. Journal of Structural and Functional Genomics, 5, 75–86. doi:10.1023/B:JSFG.0000029237.70316.52.
Johnson, E. S. (2004). Protein modification by SUMO. Annual Review of Biochemistry, 73, 355–382. doi:10.1146/annurev.biochem.73.011303.074118.
Zuo, X., Mattern, M. R., Tan, R., Li, S., Hall, J., Sterner, D. E., et al. (2005). Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein Expression and Purification, 42, 100–110. doi:10.1016/j.pep.2005.02.004.
Butt, T. R., Edavettal, S. C., Hall, J. P., & Mattern, M. R. (2005). SUMO fusion technology for difficult-to-express proteins. Protein Expression and Purification, 43, 1–9. doi:10.1016/j.pep.2005.03.016.
Luo, Y., Cho, H., Jones, R. B., Jin, C., & McKeehan, W. L. (2004). Improved production of recombinant fibroblast growth factor 7 (FGF7/KGF) from bacteria in high magnesium chloride. Protein Expression and Purification, 33, 326–331. doi:10.1016/j.pep.2003.10.013.
Laird, M. W., Cope, K., Atkinson, R., Donahoe, M., Johnson, K., & Melick, M. (2004). Keratinocyte growth factor-2 production in an ompT-deficient Escherichia coli K-12 mutant. Biotechnology Progress, 20, 44–50. doi:10.1021/bp0342147.
Wang, Y., Yuan, S., Wang, P., Liu, X., Zhan, D., & Zhang, Z. (2007). Expression, purification, and characterization of recombinant human keratinocyte growth factor-2 in Pichia pastoris. Journal of Biotechnology, 132, 44–48. doi:10.1016/j.jbiotec.2007.08.024.
Ma, Y. B., Li, Q., Xie, T. H., Li, H. J., Gang, H. Y., Dai, C. B., et al. (2001). The gene cloning, expression in E. coli and purification of human keratinocyte growth factor-2 and its bioactivity assay. Chinese Journal of Biochemistry and Molecular Biology, 17, 761–765.
Jang, J. H. (2005). Stimulation of human hair growth by the recombinant human keratinocyte growth factor-2 (KGF-2). Biotechnology Letters, 27, 749–752. doi:10.1007/s10529-005-5624-y.
Wang, J. F., Xu, D. G., Peng, S. Y., Cai, X., Zou, M. J., & Wang, J. X. (2004). Cloning and expression of recombinant human KGF-2 in E. coli. Bulletin of the Academy of Military Medical Sciences, 28, 25–27.
Acknowledgments
The work was supported by grants from Chinese National “863” High-Tech Program (2007AA02Z110), the Program of New Century Excellent Talents in University and Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents, the Science Foundation of Zhejiang Province of China (No. Z205755), Ministry of Education Incubation Foundation of Technology Innovative Project (Grant No. 706018) and Chinese National Key Basic Research Program (No. 2005CB522603).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiaoping Wu, and Changjun Nie contributed equally to the work.
Rights and permissions
About this article
Cite this article
Wu, X., Nie, C., Huang, Z. et al. Expression and Purification of Human Keratinocyte Growth Factor 2 by Fusion with SUMO. Mol Biotechnol 42, 68–74 (2009). https://doi.org/10.1007/s12033-008-9135-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-008-9135-7