Abstract
Otofaciocervical syndrome (OTFCS) is a rare genetic disorder of both autosomal recessive and autosomal dominant patterns of inheritance. It is caused by biallelic or monoallelic mutations in PAX1 or EYA1 genes, respectively. Here, we report an OTFCS2 female patient of 1st consanguineous healthy parents. She manifested facial dysmorphism, hearing loss, intellectual disability (ID), and delayed language development (DLD) as the main clinical phenotype. The novel homozygous variant c.1212dup (p.Gly405Argfs*51) in the PAX1 gene was identified by whole exome sequencing (WES), and family segregation confirmed the heterozygous status of the mutation in the parents using the Sanger sequencing. The study recorded a novel PAX1 variant representing the sixth report of OTFCS2 worldwide and the first Egyptian study expanding the geographic area where the disorder was confined.
Similar content being viewed by others
Introduction
Otofaciocervical syndrome (OTFCS) is a rare group disorder that is subclassified into two types: autosomal recessive OTFCS2 (MIM: 615,560) and autosomal dominant OTFCS1 (MIM: 166,780) due to gene mutations in PAX1 and EYA1, respectively (Patil et al. 2018). OTFCS is characterized by a combination of facial dysmorphisms, shoulder girdle anomalies, ear abnormalities accompanied by hearing loss, and intellectual disability (Pohl et al. 2013). It showed some overlapping clinical features with branchiootorenal spectrum disorders (BSDs), including branchiootic syndrome (BOS, MIM#60,258) and branchiootorenal syndrome (BORS, MIM#113,650) (Gana et al. 2019).
OTFCS2 is caused by a biallelic PAX1 mutation. The hypofunctional homozygous missense mutation c.497G < T (p.G166V) was described in the first OTFCS2 family who presented with facial dysmorphism, nasolacrimal duct and shoulder girdle abnormalities, vertebral anomalies, ear malformations, hearing loss, and mild intellectual disability (Pohl et al. 2013). However, more severe expanded features, including thymus aplasia, T-cell immunodeficiency, and recurrent infections, were observed in other successive studies (Paganini et al. 2017; Yamazaki et al. 2020; Sherlaw-Sturrock et al. 2022). To date, only fourteen affected individuals with OTFCS2 from six different families have been documented. PAX1 is a member of the paired-box (PAX) family that has already been classified into four main subfamilies, “Pax1 and Pax9; Pax2, Pax5, and Pax8; Pax3 and Pax7; Pax4 and Pax6,” according to their expression patterns, genomic organization, and the paired domain sequence. PAX1 is localized at chromosome 20p11.22, including 5 exons and encoding a 534 amino acid protein (NP_006183.2). The most conserved functional motif of PAX1 and its family is the 128 amino acid (98–226) paired-box domain (PD), which is responsible for DNA-binding activity (Blake and Ziman 2014; Thompson et al. 2021). PAX1 encodes a transcription factor protein specifically expressed during the development of the skeleton, thymus, and parathyroid glands (Wu et al. 2022). Experimentally, mice with PAX1 deficiency showed vertebral column anomalies and different degrees of thymic hypoplasia (Yamazaki et al. 2020).
Human PAX1 mutations are also involved in the Klippel-Feil (KFS) syndrome and oculo-auriculo-vertebral syndrome (OAVS) (Carter et al. 2022). The hypermethylated PAX1 promoter in esophageal squamous cell carcinoma suggests PAX1 as a tumor suppressor gene, highlighting its multifaceted roles in the human body (Nishiyama and Nakanishi 2021). Here, we present the clinical and molecular findings of the first Egyptian patient with OTFCS2 representing the sixth reported case worldwide.
Material and Methods
Patient Recruitment and Ethical Approval
A 13-year-old female patient was referred to the Multiple Congenital Anomalies (MCAs) Clinic, Center of Excellence for Human Genetics at the National Research Centre (NRC). Written informed consent was obtained from the parents according to the Declaration of Helsinki. The study was approved by the Medical Research Ethics Committee of the NRC (ID: 19,261).
Molecular Study
DNA was extracted from the peripheral blood lymphocytes using PAXgene Blood DNA (QIAGEN, Germany) according to the manufacturer’s protocol. Exome mapping was applied using the xGen Exome Research Panel v2 (Integrated DNA Technologies, Coralville, IA, USA), and sequencing was applied using the NovaSeq 6000 (Illumina, San Diego, CA, USA). Approximately 98.9% of the targeted bases were covered to a depth of ≥ 20x. All detected reads were mapped against the human reference genome (GRCh37/hg19) followed by validation of small indels and nucleotide variants using GATK best practices guidelines (Pollard et al. 2009). Variants were annotated using ANNOVAR for location and function prediction (Wang et al. 2010). Only variants deemed relevant to the patient’s clinical phenotypes were evaluated. The detected novel-related variant was examined in different databases, including ClinVar (Landrum et al. 2016), gnomAD (Karczewski and Francioli 2017), HGMD (Stenson et al. 2003), 1000 Genomes (http://browser.1000genomes.org/index.html), ExAC (http://exac.broadinstitute.org/), and GME Variome (Scott et al. 2016). The pathogenicity of the novel variant was examined by several functional prediction tools including mutation taster (Schwarz et al. 2014), CADD (Rentzsch et al. 2019), DDIG-in (Zhao et al. 2013), and TransPPMP (Nie et al. 2022). Additionally, PhyloP 100 and PhastCons 100 were checked for variant evolutionary conservation using multiple alignments based on 100 vertebrate genomes, including humans. Exon 4 of PAX1 (GenBank: NM_006192.4) was amplified utilizing standard PCR cycling conditions with specifically designed primers (F: 5′-TAATGGATGGGCACAGGACG-3′ and R: 5′-AAGTGGAGGGGACAGTCTTG-3′). The Sanger sequencing was applied using an ABI 3500 Genetic Analyser (Applied Biosystems) for variant validation and cosegregation.
Results
Clinical Results
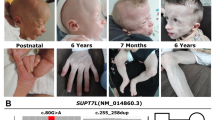
The patient was the first offspring of a 1st-degree consanguineous parent who had another healthy sibling (Fig. 1). The pregnancy history was uneventful, and the proband had a low birth weight (2 kg), as mentioned by the parents. She had a history of normal motor developmental milestones with no history of recurrent infection. A general clinical examination revealed delayed mental milestones and speech development. The patient was dysmorphic with characteristic facial features, including a long face, sparse outer third of the eyebrows, long lashes, hypertelorism, down slanting palpebral fissures, broad nose, bilateral microtia, bilateral low set ears, short neck, bilateral sloping shoulder, and toenail dystrophy, but no abnormalities were detected in the hands. Anthropometric measurements were normal according to the patient’s age (weight = 36 kg, height = 141 cm, and head circumference = 53 cm). Neurological examination showed normal muscle tone and reflexes. The auditory brainstem response (ABR) test showed bilateral sensorineural hearing loss (SNHL). Echocardiography and pelvic-abdominal ultrasound were normal. Fundoscopic and genital examinations also revealed normal findings.
a Images of the proband demonstrate a long face, sparse outer third of the eyebrows, long lashes, hypertelorism, down slanting palpebral fissures, broad nose, bilateral microtia, bilateral low set ears, short neck, bilateral sloping shoulder, and toenail dystrophy. b Family pedigree: the black arrow denotes to the affected proband
Molecular Results
A homozygous variant was identified in PAX1, one base duplication of cytosine at amino acid position 405 (NM_006192.4:c.1212dup; p.Gly405Argfs*51) in exon 4, creating a premature stop codon and truncated protein at position 456 in exon 5 of the altered sequence. The Sanger sequencing confirmed the detected homozygous variant in the proband and the carrier status in the parents, while the healthy sibling showed wild-type alleles (Fig. 2). The variant has not been detected in gnomAD, 1000 Genomes, HGMD, ClinVar, or ExAC. It has been classified as pathogenic according to the American College of Medical Genetics (ACMG) recommendations for variant classification. The variant deleterious effect has been also supported by several in silico prediction tools with relatively high conservation scores (Phylop100 = 5.324 and PhastCons 100 = 1.000) (Table 1).
Discussion
Embryonic development requires the proper regulation of various transcription factors, whose dysfunctional activity could lead to serious congenital malformations (Wu et al. 2022). PAX1 encodes a transcription factor protein that plays an essential role in different biological processes. It is especially expressed in embryogenesis during skeletal and pharyngeal pouch development, which gives rise to the thymus, tonsils, thyroid gland, and parathyroid glands (Thompson et al. 2021). Importantly, mouse point mutations in the paired-box domain of PAX1 showed a dramatic decrease in protein DNA-binding affinity, leading to skeletal deformities. Further studies declared that PAX1 deficiency is correlated with moderate thymic hypoplasia in mice, which is more exacerbated when it is accompanied by Hoxa3 haploinsufficiency (Su et al. 2001).
Human biallelic PAX1 mutation is responsible for OTFCS2 disorder, while SCID is a variable aspect among the reported patients (Pohl et al. 2013; Yamazaki et al. 2020). To date, fourteen OTFCS2 cases from six unrelated families have been reported worldwide (Table 2) (Fig. 3). Four of these families were descendants from Middle Eastern countries.
In 4 out of the 6 previously reported families, OTFCS2 was associated with the T− B+ NK+ SCID phenotype related to an underdeveloped or absent thymus. These cases had an Omenn syndrome-like phenotype, eosinophilia, and erythroderma as well as severe bacterial infections in their early life. They failed to achieve T-cell reconstitution after allogeneic hematopoietic stem cell (HPS) transplantation, leading to early death, postulating thymus transplantation as more appropriate management for OTFCS2 patients with SCID (Yamazaki et al. 2020).
Alternatively, patients of the other 2 families (n = 6) were reported to be free of SCID with no history of immunodeficiency (Pohl et al. 2013; Patil et al. 2018). Consistently, neither recurrent infections nor a history of SCID manifested in our proband (13 years old). Moreover, the patient revealed a normal complete blood picture, where both WBC and lymphocyte counts were within the normal ranges (5700 and 1208 per cubic millimeter, respectively). Normally, T cells comprise 70% of the circulating lymphocytes, so the decreased number of T cells in children with SCID usually leads to lymphopenia (Rivers and Gaspar 2015). Notably, the current patient has a reduced eosinophil count (46 per cubic millimeter), in contrast to eosinophilia reported in the T− B+ NK+ SCID phenotype.
In the current study, whole exome sequencing revealed a novel homozygous variant (NM_006192.4:c.1212dup) creating a premature stop codon in the last exon. When translation terminates either in the last exon or near the last exon–exon junction in the penultimate exon, mRNA avoids nonsense-mediated decay (NMD), possibly due to the removal of the exon junction complex (EJC) from the last exon junction (Embree et al. 2022). Therefore, residual protein activity might be attained in the reported patient associated with proper immune functioning. Hypofunctional PAX1 protein may be sufficient to stimulate the growth of the thymic epithelial lining from the third pharyngeal pouch. This might explain why some gene mutations might not be associated with congenital athymia. Therefore, we hypothesize that the activity of the mutant protein varies according to the variant type and location, which mediates whether SCID is progressed in OTFCS2 patients or not. This finding could be supported as the immune activity was similar among patients derived from the same family.
Conclusion
PAX1 mutations are generally associated with skeletal deformities that are highly comparable among all reported patients. However, the SCID is a variable aspect, where the hypofunctional PAX1 protein might be adequate to drive thymus development and activity. Molecular studies on the effects of various PAX1 variations on thymus tissue would provide useful insights into the disease genotype–phenotype correlation.
Funding
This work has been funded by the NRC grant 12060187, Egypt.
Data Availability
All study data are upon request and can be accessed by contacting the corresponding author.
References
Blake JA, Ziman MR (2014) Pax genes: regulators of lineage specification and progenitor cell maintenance. Development 141(4):737–751
Carter S, Fellows BJ, Gibson K, Bicknell LS (2022) Extending the PAX1 spectrum: a dominantly inherited variant causes oculo-auriculo-vertebral syndrome. Eur J Hum Genet 30(10):1178–1181
Embree CM, Abu-Alhasan R, Singh G (2022) Features and factors that dictate if terminating ribosomes cause or counteract nonsense-mediated mRNA decay. J Biol Chem 298(11):102592
Gana S, Valetto A, Toschi B, Sardelli I, Cappelli S, Peroni D, Bertini V (2019) Familial interstitial 6q23.2 deletion including EYA4 associated with otofaciocervical syndrome. Front Gene 10: 650
Karczewski K, Francioli L (2017) The genome aggregation database (gnomAD). MacArthur Lab 1–10
Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1):D862–D868
Nie L, Quan L, Wu T, He R, Lyu Q (2022) TransPPMP: predicting pathogenicity of frameshift and non-sense mutations by a transformer based on protein features. Bioinformatics 38(10):2705–2711
Nishiyama A, Nakanishi M (2021) Navigating the DNA methylation landscape of cancer. Trends Genet 37(11):1012–1027
Paganini I, Sestini R, Capone G, Putignano A, Contini E, Giotti I, Gensini F, Marozza A, Barilaro A, Porfirio B (2017) A novel PAX1 null homozygous mutation in autosomal recessive otofaciocervical syndrome associated with severe combined immunodeficiency. Clin Genet 92(6):664–668
Patil SJ, Das Bhowmik A, Bhat V, Satidevi Vineeth V, Vasudevamurthy R, Dalal A (2018) Autosomal recessive otofaciocervical syndrome type 2 with novel homozygous small insertion in PAX1 gene. Am J Med Genet A 176(5):1200–1206
Pohl E, Aykut A, Beleggia F, Karaca E, Durmaz B, Keupp K, Arslan E, Onay MP, Yigit G, Özkinay F (2013) A hypofunctional PAX1 mutation causes autosomal recessively inherited otofaciocervical syndrome. Hum Genet 132:1311–1320
Pollard RT, Salter I, Sanders RJ, Lucas MI, Moore CM, Mills RA, Statham PJ, Allen JT, Baker AR, Bakker DC (2009) Southern Ocean deep-water carbon export enhanced by natural iron fertilization. Nature 457(7229):577–580
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47(D1):D886–D894
Rivers L, Gaspar HB (2015) Severe combined immunodeficiency: recent developments and guidance on clinical management. Arch Dis Child 100(7):667–672
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11(4):361–362
Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, Gabriel SB, Belkadi A, Boisson B, Abel L (2016) Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet 48(9):1071–1076
Sherlaw-Sturrock C, Austin T, Baptista J, Gilmour K, Naik S (2022) Dysmorphism and immunodeficiency-one of the differential diagnoses is PAX1 related otofaciocervical syndrome type 2. Eur J Med Genet 65(7):104523
Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN (2003) Human Gene Mutation Database (HGMD®): 2003 update. Hum Mutat 21(6):577–581
Su D-m, Ellis S, Napier A, Lee K, Manley NR (2001) Hoxa3 and pax1 regulate epithelial cell death and proliferation during thymus and parathyroid organogenesis. Dev Biol 236(2):316–329
Thompson B, Davidson EA, Liu W, Nebert DW, Bruford EA, Zhao H, Dermitzakis ET, Thompson DC, Vasiliou V (2021) Overview of PAX gene family: analysis of human tissue-specific variant expression and involvement in human disease. Hum Genet 140:381–400
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variations from next-generation sequencing data. Nucl Acids Res 38:e164
Wu W, Kong X, Jia Y, Jia Y, Ou W, Dai C, Li G, Gao R (2022) An overview of PAX1: expression, function and regulation in development and diseases. Front Cell Dev Biol 10
Yamazaki Y, Urrutia R, Franco LM, Giliani S, Zhang K, Alazami AM, Dobbs AK, Masneri S, Joshi A, Otaizo-Carrasquero F (2020) PAX1 is essential for development and function of the human thymus. Sci Immunol 5(44): eaax1036
Zhao H, Yang Y, Lin H, Zhang X, Mort M, Cooper DN, Liu Y, Zhou Y (2013) DDIG-in: discriminating between disease-associated and neutral non-frameshifting micro-indels. Genome Biol 14:1–13
Acknowledgements
We are appreciative to the parents who agreed to take part in the study and donate samples.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Nagham M. Elbagoury, Asmaa F. Abdel-Aleem, Wessam E. Sharaf-Eldin, Engy A. Ashaat, and Mona L. Esswai contributed to the study conception and design. Engy A. Ashaat. investigated the case. Nagham M. Elbagoury, Wessam E. Sharaf-Eldin, and Asmaa F. Abdel-Aleem. carried out molecular analysis Asmaa F. Abdel-Aleem wrote the first draft of the manuscript. Wessam E. Sharaf-Eldin, Nagham M. Elbagoury, Mona L. Esswai, and Engy A. Ashaat revised the manuscript.
Corresponding author
Ethics declarations
Declarations
Helsinki’s guiding principles were followed during the study. The study has been granted approval by the NRC Medical Research Ethics Committee (ID: 19261). After fully explaining the study to parents, their written agreement was acquired.
Competing Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elbagoury, N.M., Abdel-Aleem, A.F., Sharaf-Eldin, W.E. et al. A Novel Truncating Mutation in PAX1 Gene Causes Otofaciocervical Syndrome Without Immunodeficiency. J Mol Neurosci 73, 976–982 (2023). https://doi.org/10.1007/s12031-023-02170-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-023-02170-7