Abstract
Nucleotide-binding and oligomerization domain-like receptors (NLRs) are central regulators of pathogen recognition, the induction of innate immune effectors and inflammation with utmost importance in human diseases such as inflammatory bowel diseases. Most NLRs are key mediators of inflammasome complexes that activate caspase-1 and drive proteolytic processing of pro-inflammatory cytokines; however, a few tightly regulate inflammasome-independent activation of nuclear factor-κB and mitogen-activated protein kinase pathways. NLR signaling has evolved in intestinal epithelial cells to avoid overactive inflammatory responses toward the resident microbiota and to preserve epithelial barrier integrity and functions by maintaining homeostasis. In the present review, I examine new insights into the role of the NLRs in antimicrobial defenses. I pay particular attention to the emerging role of these receptors in engaging a complex cross talk between cell death and innate immunity pathways. Furthermore, I discuss the physiological functions of the NLRs in shaping the innate immune response within the intestine, maintaining homeostasis, inducing tissue repair following injury and promoting tumorigenesis.



Similar content being viewed by others
References
Klein J. Self-nonself discrimination, histoincompatibility, and the concept of immunology. Immunogenetics. 1999;50(3–4):116–23.
Rescigno M. Before they were gut dendritic cells. Immunity. 2009;31(3):454–6.
Hand T, Belkaid Y. Microbial control of regulatory and effector T cell responses in the gut. Curr Opin Immunol. 2010;22(1):63–72.
Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228–31.
Xu J, Gordon JI. Honor thy symbionts. Proc Nat Acad Sci USA. 2003;100(18):10452–9.
Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4(12):953–64.
Bjerknes M, Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006;419:337–83.
Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140(6):859–70.
Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5.
Yeretssian G, Labbe K, Saleh M. Molecular regulation of inflammation and cell death. Cytokine. 2008;43(3):380–90.
Chen G, et al. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–98.
Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11(1):9–20.
Inohara N, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-κB. J Biol Chem. 1999;274(21):14560–7.
Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr Opin Plant Biol. 2004;7(4):391–9.
Harton JA, et al. Cutting edge: CATERPILLER—a large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169(8):4088–93.
Ting JP, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28(3):285–7.
Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–72.
Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278(8):5509–12.
Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–7.
Girardin SE, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300(5625):1584–7.
Benko S, Philpott DJ, Girardin SE. The microbial and danger signals that activate Nod-like receptors. Cytokine. 2008;43(3):368–73.
Bertrand MJ, et al. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30(6):789–801.
Abbott DW, et al. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14(24):2217–27.
Krieg A, et al. XIAP mediates NOD signaling via interaction with RIP2. Proc Nat Acad Sci USA. 2009;106(34):14524–9.
Abbott DW, et al. Coordinated regulation of toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27(17):6012–25.
Hasegawa M, et al. A critical role of RICK/RIP2 polyubiquitination in nod-induced NF-κB activation. EMBO J. 2008;27(2):373–83.
Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nat Rev Mol Cell Biol. 2007;8(1):49–62.
Hsu YM, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8(2):198–205.
LeBlanc PM, et al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe. 2008;3(3):146–57.
Fritz JH, et al. Synergistic stimulation of human monocytes and dendritic cells by toll-like receptor 4 and NOD1- and NOD2-activating agonists. Eur J Immunol. 2005;35(8):2459–70.
Viala J, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5(11):1166–74.
Watanabe T, et al. NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Investig. 2010;120(5):1645–62.
Hasegawa M, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J Immunol. 2011;186(8):4872–80.
Berrington WR, et al. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. Eur J Immunol. 2010;40(12):3519–27.
Frutuoso MS, et al. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect/Inst Pasteur. 2010;12(11):819–27.
Boneca IG, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Nat Acad Sci USA. 2007;104(3):997–1002.
Travassos LH, et al. Toll-like receptor 2-dependent bacterial sensing does not occur via peptidoglycan recognition. EMBO Rep. 2004;5(10):1000–6.
Travassos LH, et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280(44):36714–8.
Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307(5710):731–4.
Deshmukh HS, et al. Critical role of NOD2 in regulating the immune response to Staphylococcus aureus. Infect Immun. 2009;77(4):1376–82.
Shimada K, et al. The NOD/RIP2 pathway is essential for host defenses against Chlamydophila pneumoniae lung infection. PLoS Pathog. 2009;5(4):e1000379.
Opitz B, et al. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279(35):36426–32.
Kim YG, et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34(5):769–80.
Steimle V, et al. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265(5168):106–9.
Nickerson K, et al. Dendritic cell-specific MHC class II transactivator contains a caspase recruitment domain that confers potent transactivation activity. J Biol Chem. 2001;276(22):19089–93.
LeibundGut-Landmann S, et al. Mini-review: specificity and expression of CIITA, the master regulator of MHC class II genes. Eur J Immunol. 2004;34(6):1513–25.
Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451(7178):573–7.
Tattoli I, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-κB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9(3):293–300.
Xia X, et al. NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843–53.
Allen IC, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-κB signaling pathways. Immunity. 2011;34(6):854–65.
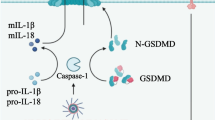
Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–26.
Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–32.
Shao W, et al. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem. 2007;282(50):36321–9.
Bruey JM, et al. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-κB and caspase-1 activation in macrophages. J Biol Chem. 2004;279(50):51897–907.
Grenier JM, et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-κB and caspase-1. FEBS Lett. 2002;530(1–3):73–8.
Wang L, et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-κB and caspase-1-dependent cytokine processing. J Biol Chem. 2002;277(33):29874–80.
Arthur JC, et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J Immunol. 2010;185(8):4515–9.
Zaki MH, et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20(5):649–60.
Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–57.
Faustin B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25(5):713–24.
Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38(2):240–4.
Hsu LC, et al. A NOD2–NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Nat Acad Sci USA. 2008;105(22):7803–8.
Jin Y, et al. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356(12):1216–25.
Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440(7081):228–32.
McNeela EA, et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog. 2010;6(11):e1001191.
Harder J, et al. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-κB activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol. 2009;183(9):5823–9.
Craven RR, et al. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS One. 2009;4(10):e7446.
Gross O, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459(7245):433–6.
Hise AG, et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5(5):487–97.
Kumar H, et al. Involvement of the NLRP3 inflammasome in innate and humoral adaptive immune responses to fungal beta-glucan. J Immunol. 2009;183(12):8061–7.
Kanneganti TD, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440(7081):233–6.
Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452(7183):103–7.
Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–75.
Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–65.
Ichinohe T, et al. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206(1):79–87.
Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11(5):404–10.
Kahlenberg JM, Dubyak GR. Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+ release. Am J Physiol Cell Physiol. 2004;286(5):C1100–8.
Kanneganti TD, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of toll-like receptor signaling. Immunity. 2007;26(4):433–43.
Martinon F, et al. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440(7081):237–41.
Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320(5876):674–7.
Cassel SL, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Nat Acad Sci USA. 2008;105(26):9035–40.
Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847–56.
Eisenbarth SC, et al. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453(7198):1122–6.
Halle A, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9(8):857–65.
Petrilli V, et al. The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol. 2007;19(6):615–22.
Cruz CM, et al. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282(5):2871–9.
Zhou R, et al. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11(2):136–40.
Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–8.
Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3(8):e111.
Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7(3):318–25.
Amer A, et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281(46):35217–23.
Brodsky IE, et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7(5):376–87.
Franchi L, et al. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37(11):3030–9.
Miao EA, et al. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Nat Acad Sci USA. 2008;105(7):2562–7.
Sutterwala FS, et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204(13):3235–45.
Poyet JL, et al. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276(30):28309–13.
Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65.
Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9(10):1171–8.
Fortier A, et al. Restriction of Legionella pneumophila replication in macrophages requires concerted action of the transcriptional regulators Irf1 and Irf8 and nod-like receptors Naip5 and Nlrc4. Infect Immun. 2009;77(11):4794–805.
Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477(7366):592–5.
Zhao Y, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477(7366):596–600.
Stehlik C, Dorfleutner A. COPs and POPs: modulators of inflammasome activity. J Immunol. 2007;179(12):7993–8.
Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117(5):561–74.
Humke EW, et al. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000;103(1):99–111.
Druilhe A, et al. Regulation of IL-1beta generation by pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8(6):649–57.
Saleh M, et al. Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature. 2006;440(7087):1064–8.
Fischer H, et al. Human caspase 12 has acquired deleterious mutations. Biochem Biophys Res Commun. 2002;293(2):722–6.
Yeretssian G, et al. Gender differences in expression of the human caspase-12 long variant determines susceptibility to Listeria monocytogenes infection. Proc Nat Acad Sci USA. 2009;106(22):9016–20.
Saleh M, et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429(6987):75–9.
Xue Y, et al. Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78(4):659–70.
Labbe K, et al. Caspase-12 dampens the immune response to malaria independently of the inflammasome by targeting NF-κB signaling. J Immunol. 2010;185(9):5495–502.
Razmara M, et al. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277(16):13952–8.
Rosenstiel P, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Nat Acad Sci USA. 2006;103(9):3280–5.
Fairbrother WJ, et al. The PYRIN domain: a member of the death domain-fold superfamily. Prot Sci Publ Prot Soc. 2001;10(9):1911–8.
Stehlik C, et al. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-κB and pro-caspase-1 regulation. Biochem J. 2003;373(Pt 1):101–13.
Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-κB and disrupts ASC: CLR interactions. J Immunol. 2007;178(6):3837–45.
Dorfleutner A, et al. A Shope fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35(3):685–94.
Guarda G, et al. T cells dampen innate immune responses through inhibition of NLRP1 and NLRP3 inflammasomes. Nature. 2009;460(7252):269–73.
Cui J, et al. NLRC5 negatively regulates the NF-κB and type I interferon signaling pathways. Cell. 2010;141(3):483–96.
Benko S, et al. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185(3):1681–91.
Bruey JM, et al. Bcl-2 and Bcl-XL regulate proinflammatory caspase-1 activation by interaction with NALP1. Cell. 2007;129(1):45–56.
Greten FR, et al. NF-κB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130(5):918–31.
Opferman JT. Apoptosis in the development of the immune system. Cell Death Differ. 2008;15(2):234–42.
Henson PM. Dampening inflammation. Nat Immunol. 2005;6(12):1179–81.
Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8(5):405–13.
Nakahira K, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–30.
Zhou R, et al. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469(7329):221–5.
Bauernfeind F, et al. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol. 2011;187(2):613–7.
Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16(1):90–7.
Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11(1):55–62.
Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–8.
Cain K, et al. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J Biol Chem. 2001;276(45):41985–90.
Brown GC, Borutaite V. Regulation of apoptosis by the redox state of cytochrome c. Biochim Biophys Acta. 2008;1777(7–8):877–81.
Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene. 2008;27(50):6434–51.
Yuan XM, et al. Lysosomal destabilization in p53-induced apoptosis. Proc Nat Acad Sci USA. 2002;99(9):6286–91.
Green DR. At the gates of death. Cancer Cell. 2006;9(5):328–30.
Faustin B, et al. Mechanism of Bcl-2 and Bcl-X(L) inhibition of NLRP1 inflammasome: loop domain-dependent suppression of ATP binding and oligomerization. Proc Nat Acad Sci USA. 2009;106(10):3935–40.
Yeretssian G, et al. Non-apoptotic role of BID in inflammation and innate immunity. Nature. 2011;474(7349):96–9.
Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621.
Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24(1):4–10.
Bouskra D, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–10.
Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–18.
Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4(6):478–85.
Haverson K, et al. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol. 2007;119(3–4):243–53.
Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328(5986):1705–9.
Dong C. Diversification of T-helper-cell lineages: finding the family root of IL-17-producing cells. Nat Rev Immunol. 2006;6(4):329–33.
Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41.
Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98.
Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5.
Petnicki-Ocwieja T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Nat Acad Sci USA. 2009;106(37):15813–8.
Rehman A, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60(10):1354–62.
Hirota SA, et al. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm Bowel Dis. 2011;17(6):1359–72.
Villani AC, et al. Common variants in the NLRP3 region contribute to Crohn’s disease susceptibility. Nat Genet. 2009;41(1):71–6.
McGovern DP, et al. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14(10):1245–50.
Lu WG, et al. Association of NOD1 (CARD4) insertion/deletion polymorphism with susceptibility to IBD: a meta-analysis. World J Gastroenterol. 2010;16(34):4348–56.
Hasegawa M, et al. Differential release and distribution of Nod1 and Nod2 immunostimulatory molecules among bacterial species and environments. J Biol Chem. 2006;281(39):29054–63.
Chen GY, et al. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68(24):10060–7.
Dupaul-Chicoine J, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32(3):367–78.
Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32(3):379–91.
Allen IC, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207(5):1045–56.
Salcedo R, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207(8):1625–36.
Zaki MH, et al. IL-18 production downstream of the Nlrp3 inflammasome confers protection against colorectal tumor formation. J Immunol. 2010;185(8):4912–20.
Bauer C, et al. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010;59(9):1192–9.
Hu B, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. Proc Nat Acad Sci USA. 2010;107(50):21635–40.
Chen GY, et al. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186(12):7187–94.
Normand S, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Nat Acad Sci USA. 2011;108(23):9601–6.
Acknowledgments
Garabet Yeretssian thanks his laboratory members for reading and commenting on this manuscript and N. Fodil-Cornu for critical reading of the manuscript, comments, and discussions. Work in Garabet Yeretssian’s laboratory is supported by the Helmsley Foundation. The author declares no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yeretssian, G. Effector functions of NLRs in the intestine: innate sensing, cell death, and disease. Immunol Res 54, 25–36 (2012). https://doi.org/10.1007/s12026-012-8317-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-012-8317-3