Abstract
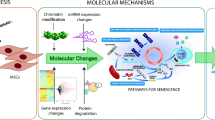
Clinical trials using human mesenchymal stem/stromal cells (hMSCs) for cell replacement therapy showed varied outcomes, where cells’ efficacy has been perceived as the limiting factor. In particular, the quality and number of the expanded cells in vitro. In this study, we aimed to determine molecular signatures of hMSCs derived from the pulp of extracted deciduous teeth (SHED) and Wharton’s jelly (WJSCs) that associated with cellular ageing during in vitro passaging. We observed distinct phenotypic changes resembling proliferation reduction, cell enlargement, an increase cell population in G2/M phase, and differentially expressed of tumor suppressor p53 in passage (P) 6 as compared to P3, which indicating in vitro cell senescence. The subsequent molecular analysis showed a set of diverse differentially expressed miRNAs and mRNAs involved in maintaining cell proliferation and stemness properties. Considering the signaling pathway related to G2/M DNA damage regulation is widely recognized as part of anti-proliferation mechanism controlled by p53, we explored possible miRNA-mRNA interaction in this regulatory pathway based on genomic coordinates retrieved from miRanda. Our work reveals the potential reason for SHED underwent proliferation arrest due to the direct impinge on the expression of CKS1 by miRNAs specifically miR-22 and miR-485-5p which lead to down regulation of CDK1 and Cyclin B. It is intended that our study will contribute to the understanding of these miRNA/mRNA driving the biological process and regulating different stages of cell cycle is beneficial in developing effective rejuvenation strategies in order to obtain quality stem cells for transplantation.






Similar content being viewed by others
Change history
17 July 2020
The original version of this article unfortunately contained a mistake.
References
Estrada, J. C., Torres, Y., Benguria, A., Dopazo, A., Roche, E., Carrera-Quintanar, L., Perez, R. A., Enriquez, J. A., Torres, R., Ramirez, J. C., Samper, E., & Bernad, A. (2013). Human mesenchymal stem cell-replicative senescence and oxidative stress are closely linked to aneuploidy. Cell Death & Disease, 4, e691. https://doi.org/10.1038/cddis.2013.211.
Sepulveda, J. C., Tome, M., Fernandez, M. E., Delgado, M., Campisi, J., Bernad, A., & Gonzalez, M. A. (2014). Cell senescence abrogates the therapeutic potential of human mesenchymal stem cells in the lethal endotoxemia model. Stem Cells (Dayton, Ohio), 32, 1865–1877. https://doi.org/10.1002/stem.1654.
Wagner, W., Bork, S., Lepperdinger, G., Joussen, S., Ma, N., Strunk, D., & Koch, C. (2010). How to track cellular aging of mesenchymal stromal cells? Aging, 2, 224–230. https://doi.org/10.18632/aging.100136.
Wagner, W., Horn, P., Castoldi, M., Diehlmann, A., Bork, S., Saffrich, R., Benes, V., Blake, J., Pfister, S., Eckstein, V., & Ho, A. D. (2008). Replicative senescence of mesenchymal stem cells: A continuous and organized process. PLoS One, 3, e2213. https://doi.org/10.1371/journal.pone.0002213.
Bentivegna, A., Roversi, G., Riva, G., Paoletta, L., Redaelli, S., Miloso, M., Tredici, G., & Dalpra, L. (2016). The effect of culture on human bone marrow Mesenchymal stem cells: Focus on DNA methylation profiles. Stem Cells International, 2016, 5656701. https://doi.org/10.1155/2016/5656701.
Legzdina, D., Romanauska, A., Nikulshin, S., Kozlovska, T., & Berzins, U. (2016). Characterization of senescence of culture-expanded human adipose-derived Mesenchymal stem cells. International Journal of Stem Cells, 9, 124–136. https://doi.org/10.15283/ijsc.2016.9.1.124.
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
Shomron, N., & Levy, C. (2009). MicroRNA-biogenesis and pre-mRNA splicing crosstalk. Journal of Biomedicine & Biotechnology, 2009, 594678. https://doi.org/10.1155/2009/594678.
Lee, J., Li, Z., Brower-Sinning, R., & John, B. (2007). Regulatory circuit of human microRNA biogenesis. PLoS Computational Biology, 3, e67. https://doi.org/10.1371/journal.pcbi.0030067.
Benhamed, M., Herbig, U., Ye, T., Dejean, A., & Bischof, O. (2012). Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nature Cell Biology, 14, 266–275. https://doi.org/10.1038/ncb2443.
Dhahbi, J. M., Atamna, H., Boffelli, D., Magis, W., Spindler, S. R., & Martin, D. I. (2011). Deep sequencing reveals novel microRNAs and regulation of microRNA expression during cell senescence. PLoS One, 6, e20509. https://doi.org/10.1371/journal.pone.0020509.
Liu, F. J., Wen, T., & Liu, L. (2012). MicroRNAs as a novel cellular senescence regulator. Ageing Research Reviews, 11, 41–50. https://doi.org/10.1016/j.arr.2011.06.001.
Govindasamy, V., Abdullah, A. N., Ronald, V. S., Musa, S., Ab Aziz, Z. A., Zain, R. B., Totey, S., Bhonde, R. R., & Abu Kasim, N. H. (2010). Inherent differential propensity of dental pulp stem cells derived from human deciduous and permanent teeth. Journal of Endodontics, 36, 1504–1515. https://doi.org/10.1016/j.joen.2010.05.006.
Koch, C. M., Reck, K., Shao, K., Lin, Q., Joussen, S., Ziegler, P., Walenda, G., Drescher, W., Opalka, B., May, T., Brummendorf, T., Zenke, M., Saric, T., & Wagner, W. (2013). Pluripotent stem cells escape from senescence-associated DNA methylation changes. Genome Research, 23, 248–259. https://doi.org/10.1101/gr.141945.112.
Brown, P. T., Squire, M. W., & Li, W. J. (2014). Characterization and evaluation of mesenchymal stem cells derived from human embryonic stem cells and bone marrow. Cell and Tissue Research, 358, 149–164. https://doi.org/10.1007/s00441-014-1926-5.
Wegmeyer, H., Broske, A. M., Leddin, M., Kuentzer, K., Nisslbeck, A. K., Hupfeld, J., Wiechmann, K., Kuhlen, J., von Schwerin, C., Stein, C., Knothe, S., Funk, J., Huss, R., & Neubauer, M. (2013). Mesenchymal stromal cell characteristics vary depending on their origin. Stem Cells and Development, 22, 2606–2618. https://doi.org/10.1089/scd.2013.0016.
Hong, H., Takahashi, K., Ichisaka, T., Aoi, T., Kanagawa, O., Nakagawa, M., Okita, K., & Yamanaka, S. (2009). Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature, 460, 1132–1135. https://doi.org/10.1038/nature08235.
Kawamura, T., Suzuki, J., Wang, Y. V., Menendez, S., Morera, L. B., Raya, A., Wahl, G. M., & Izpisua Belmonte, J. C. (2009). Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature, 460, 1140–1144. https://doi.org/10.1038/nature08311.
Solozobova, V., & Blattner, C. (2011). p53 in stem cells. World Journal of Biological Chemistry, 2, 202–214. https://doi.org/10.4331/wjbc.v2.i9.202.
Gu, Y., Li, T., Ding, Y., Sun, L., Tu, T., Zhu, W., Hu, J., & Sun, X. (2016). Changes in mesenchymal stem cells following long-term culture in vitro. Molecular Medicine Reports, 13, 5207–5215. https://doi.org/10.3892/mmr.2016.5169.
Mandal, P. K., Blanpain, C., & Rossi, D. J. (2011). DNA damage response in adult stem cells: Pathways and consequences. Nature reviews. Molecular Cell Biology, 12, 198–202. https://doi.org/10.1038/nrm3060.
Simara, P., Tesarova, L., Rehakova, D., Matula, P., Stejskal, S., Hampl, A., & Koutna, I. (2017). DNA double-strand breaks in human induced pluripotent stem cell reprogramming and long-term in vitro culturing. Stem Cell Research & Therapy, 8, 73. https://doi.org/10.1186/s13287-017-0522-5.
Whitfield, M. J., Lee, W. C., & Van Vliet, K. J. (2013). Onset of heterogeneity in culture-expanded bone marrow stromal cells. Stem Cell Research, 11, 1365–1377. https://doi.org/10.1016/j.scr.2013.09.004.
Muraglia, A., Cancedda, R., & Quarto, R. (2000). Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. Journal of Cell Science, 113(Pt 7), 1161–1166.
Aponte, P. M., & Caicedo, A. (2017). Stemness in Cancer: Stem cells, Cancer stem cells, and their microenvironment. Stem Cells International, 2017, 5619472. https://doi.org/10.1155/2017/5619472.
Viatour, P. (2012). Bridges between cell cycle regulation and self-renewal maintenance. Genes & Cancer, 3, 670–677. https://doi.org/10.1177/1947601913481355.
Ba, H., Wang, D., & Li, C. (2016). MicroRNA profiling of antler stem cells in potentiated and dormant states and their potential roles in antler regeneration. Molecular Genetics and Genomics : MGG, 291, 943–955. https://doi.org/10.1007/s00438-015-1158-8.
Filipowicz, W., Bhattacharyya, S. N., & Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nature Reviews. Genetics, 9, 102–114. https://doi.org/10.1038/nrg2290.
Heinrich, E. M., & Dimmeler, S. (2012). MicroRNAs and stem cells: Control of pluripotency, reprogramming, and lineage commitment. Circulation Research, 110, 1014–1022. https://doi.org/10.1161/circresaha.111.243394.
Maroney, P. A., Yu, Y., Fisher, J., & Nilsen, T. W. (2006). Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nature Structural & Molecular Biology, 13, 1102–1107. https://doi.org/10.1038/nsmb1174.
Thomson, J. M., Newman, M., Parker, J. S., Morin-Kensicki, E. M., Wright, T., & Hammond, S. M. (2006). Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Development, 20, 2202–2207. https://doi.org/10.1101/gad.1444406.
Zhao, B., Yang, D., Jiang, J., Li, J., Fan, C., Huang, M., Fan, Y., Jin, Y., & Jin, Y. (2014). Genome-wide mapping of miRNAs expressed in embryonic stem cells and pluripotent stem cells generated by different reprogramming strategies. BMC Genomics, 15, 488. https://doi.org/10.1186/1471-2164-15-488.
Guo, L., Zhao, R. C., & Wu, Y. (2011). The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Experimental Hematology, 39, 608–616. https://doi.org/10.1016/j.exphem.2011.01.011.
Meng, X., Sun, B., Xue, M., Xu, P., Hu, F., & Xiao, Z. (2016). Comparative analysis of microRNA expression in human mesenchymal stem cells from umbilical cord and cord blood. Genomics, 107, 124–131. https://doi.org/10.1016/j.ygeno.2016.02.006.
Ren, J., Jin, P., Wang, E., Marincola, F. M., & Stroncek, D. F. (2009). MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. Journal of Translational Medicine, 7, 20. https://doi.org/10.1186/1479-5876-7-20.
Sharma, A., & Wu, J. C. (2013). MicroRNA expression profiling of human-induced pluripotent and embryonic stem cells. Methods in Molecular Biology (Clifton, N.J.), 936, 247–256. https://doi.org/10.1007/978-1-62703-083-0_19.
Tang, F., Hajkova, P., Barton, S. C., Lao, K., & Surani, M. A. (2006). MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Research, 34, e9. https://doi.org/10.1093/nar/gnj009.
Dalton, S. (2013). Signaling networks in human pluripotent stem cells. Current Opinion in Cell Biology, 25, 241–246. https://doi.org/10.1016/j.ceb.2012.09.005.
Ito, K., & Suda, T. (2014). Metabolic requirements for the maintenance of self-renewing stem cells. Nature reviews. Molecular Cell Biology, 15, 243–256. https://doi.org/10.1038/nrm3772.
Qi, Y., Liu, J., Saadat, S., Tian, X., Han, Y., Fong, G. H., Pandolfi, P. P., Lee, L. Y., & Li, S. (2015). PTEN induces apoptosis and cavitation via HIF-2-dependent Bnip3 upregulation during epithelial lumen formation. Cell Death and Differentiation, 22, 875–884. https://doi.org/10.1038/cdd.2014.185.
Takase, O., Yoshikawa, M., Idei, M., Hirahashi, J., Fujita, T., Takato, T., Isagawa, T., Nagae, G., Suemori, H., Aburatani, H., & Hishikawa, K. (2013). The role of NF-kappaB signaling in the maintenance of pluripotency of human induced pluripotent stem cells. PLoS One, 8, e56399. https://doi.org/10.1371/journal.pone.0056399.
Ming, M., & He, Y. Y. (2012). PTEN in DNA damage repair. Cancer Letters, 319, 125–129. https://doi.org/10.1016/j.canlet.2012.01.003.
Shen, W. H., Balajee, A. S., Wang, J., Wu, H., Eng, C., Pandolfi, P. P., & Yin, Y. (2007). Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell, 128, 157–170. https://doi.org/10.1016/j.cell.2006.11.042.
Chien, Y., Scuoppo, C., Wang, X., Fang, X., Balgley, B., Bolden, J. E., Premsrirut, P., Luo, W., Chicas, A., Lee, C. S., Kogan, S. C., & Lowe, S. W. (2011). Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes & Development, 25, 2125–2136. https://doi.org/10.1101/gad.17276711.
Hermeking, H., & Benzinger, A. (2006). 14-3-3 proteins in cell cycle regulation. Seminars in Cancer Biology, 16, 183–192. https://doi.org/10.1016/j.semcancer.2006.03.002.
Morrison, D. K. (2009). The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends in Cell Biology, 19, 16–23. https://doi.org/10.1016/j.tcb.2008.10.003.
Harries, L. W., Fellows, A. D., Pilling, L. C., Hernandez, D., Singleton, A., Bandinelli, S., Guralnik, J., Powell, J., Ferrucci, L., & Melzer, D. (2012). Advancing age is associated with gene expression changes resembling mTOR inhibition: Evidence from two human populations. Mechanisms of Ageing and Development, 133, 556–562. https://doi.org/10.1016/j.mad.2012.07.003.
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M., & Kim, D. H. (2010). mTOR regulation of autophagy. FEBS Letters, 584, 1287–1295. https://doi.org/10.1016/j.febslet.2010.01.017.
Inoki, K., Zhu, T., & Guan, K. L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell, 115, 577–590.
Shaltiel, I. A., Krenning, L., Bruinsma, W., & Medema, R. H. (2015). The same, only different - DNA damage checkpoints and their reversal throughout the cell cycle. Journal of Cell Science, 128, 607–620. https://doi.org/10.1242/jcs.163766.
Schwermer, M., Lee, S., Koster, J., van Maerken, T., Stephan, H., Eggert, A., Morik, K., Schulte, J. H., & Schramm, A. (2015). Sensitivity to cdk1-inhibition is modulated by p53 status in preclinical models of embryonal tumors. Oncotarget, 6, 15425–15435. https://doi.org/10.18632/oncotarget.3908.
Branzei, D., & Foiani, M. (2008). Regulation of DNA repair throughout the cell cycle. Nature reviews. Molecular Cell Biology, 9, 297–308. https://doi.org/10.1038/nrm2351.
Rhind, N., & Russell, P. (2012). Signaling pathways that regulate cell division. Cold Spring Harbor Perspectives in Biology, 4. https://doi.org/10.1101/cshperspect.a005942.
Westbrook, L., Manuvakhova, M., Kern, F. G., Estes, N. R., 2nd, Ramanathan, H. N., & Thottassery, J. V. (2007). Cks1 regulates cdk1 expression: A novel role during mitotic entry in breast cancer cells. Cancer Research, 67, 11393–11401. https://doi.org/10.1158/0008-5472.can-06-4173.
Satyanarayana, A., Berthet, C., Lopez-Molina, J., Coppola, V., Tessarollo, L., & Kaldis, P. (2008). Genetic substitution of Cdk1 by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2. Development (Cambridge, England), 135, 3389–3400. https://doi.org/10.1242/dev.024919.
Tsai, Y. S., Chang, H. C., Chuang, L. Y., & Hung, W. C. (2005). RNA silencing of Cks1 induced G2/M arrest and apoptosis in human lung cancer cells. IUBMB Life, 57, 583–589. https://doi.org/10.1080/15216540500215531.
Chen, H., Lu, Q., Fei, X., Shen, L., Jiang, D., & Dai, D. (2016). miR-22 inhibits the proliferation, motility, and invasion of human glioblastoma cells by directly targeting SIRT1. Tumour biology : The Journal of The International Society for Oncodevelopmental Biology and Medicine, 37, 6761–6768. https://doi.org/10.1007/s13277-015-4575-8.
Lou, C., Xiao, M., Cheng, S., Lu, X., Jia, S., Ren, Y., & Li, Z. (2016). MiR-485-3p and miR-485-5p suppress breast cancer cell metastasis by inhibiting PGC-1alpha expression. Cell Death & Disease, 7, e2159. https://doi.org/10.1038/cddis.2016.27.
Wakeling, L. A., Ions, L. J., & Ford, D. (2009). Could Sirt1-mediated epigenetic effects contribute to the longevity response to dietary restriction and be mimicked by other dietary interventions? Age (Dordrecht, Netherlands), 31, 327–341. https://doi.org/10.1007/s11357-009-9104-5.
Luo, J., Nikolaev, A. Y., Imai, S., Chen, D., Su, F., Shiloh, A., Guarente, L., & Gu, W. (2001). Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell, 107, 137–148.
Ong, A. L. C., & Ramasamy, T. S. (2018). Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Research Reviews, 43, 64–80. https://doi.org/10.1016/j.arr.2018.02.004.
Yeung, F., Hoberg, J. E., Ramsey, C. S., Keller, M. D., Jones, D. R., Frye, R. A., & Mayo, M. W. (2004). Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. The EMBO Journal, 23, 2369–2380. https://doi.org/10.1038/sj.emboj.7600244.
Nemoto, S., Fergusson, M. M., & Finkel, T. (2005). SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. The Journal of Biological Chemistry, 280, 16456–16460. https://doi.org/10.1074/jbc.M501485200.
Faustman, D. L., & Davis, M. (2013). TNF receptor 2 and disease: Autoimmunity and regenerative medicine. Frontiers in Immunology, 4, 478. https://doi.org/10.3389/fimmu.2013.00478.
Naude, P. J., den Boer, J. A., Luiten, P. G., & Eisel, U. L. (2011). Tumor necrosis factor receptor cross-talk. The FEBS Journal, 278, 888–898. https://doi.org/10.1111/j.1742-4658.2011.08017.x.
Rauert, H., Stuhmer, T., Bargou, R., Wajant, H., & Siegmund, D. (2011). TNFR1 and TNFR2 regulate the extrinsic apoptotic pathway in myeloma cells by multiple mechanisms. Cell Death & Disease, 2, e194. https://doi.org/10.1038/cddis.2011.78.
Tuncer, S., & Banerjee, S. (2015). Eicosanoid pathway in colorectal cancer: Recent updates. World Journal of Gastroenterology, 21, 11748–11766. https://doi.org/10.3748/wjg.v21.i41.11748.
Mishra, K. P., Jain, S., Ganju, L., & Singh, S. B. (2014). Hypoxic stress induced TREM-1 and inflammatory chemokines in human peripheral blood mononuclear cells. Indian Journal of Clinical Biochemistry : IJCB, 29, 133–138. https://doi.org/10.1007/s12291-013-0345-9.
Hichor, M., Sundaram, V. K., Eid, S. A., Abdel-Rassoul, R., Petit, P. X., Borderie, D., Bastin, J., Eid, A. A., & Manuel, M. (2018). Liver X Receptor exerts a protective effect against the oxidative stress in the peripheral nerve. Liver X Receptor exerts a protective effect against the oxidative stress in the peripheral nerve., 8, 2524. https://doi.org/10.1038/s41598-018-20980-3.
Lukin, D. J., Carvajal, L. A., Liu, W. J., Resnick-Silverman, L., & Manfredi, J. J. (2015). p53 promotes cell survival due to the reversibility of its cell-cycle checkpoints. Molecular cancer research : MCR, 13, 16–28. https://doi.org/10.1158/1541-7786.mcr-14-0177.
Lee, J. H., Park, S. J., Jeong, S. Y., Kim, M. J., Jun, S., Lee, H. S., Chang, I. Y., Lim, S. C., Yoon, S. P., Yong, J., & You, H. J. (2015). MicroRNA-22 suppresses DNA repair and promotes genomic instability through targeting of MDC1. Cancer Research, 75, 1298–1310. https://doi.org/10.1158/0008-5472.can-14-2783.
Huang, Z. P., Chen, J., Seok, H. Y., Zhang, Z., Kataoka, M., Hu, X., & Wang, D. Z. (2013). MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circulation Research, 112, 1234–1243. https://doi.org/10.1161/circresaha.112.300682.
Wang, Z., Qin, G., & Zhao, T. C. (2014). HDAC4: Mechanism of regulation and biological functions. Epigenomics, 6, 139–150. https://doi.org/10.2217/epi.13.73.
Xu, D., Takeshita, F., Hino, Y., Fukunaga, S., Kudo, Y., Tamaki, A., Matsunaga, J., Takahashi, R. U., Takata, T., Shimamoto, A., Ochiya, T., & Tahara, H. (2011). miR-22 represses cancer progression by inducing cellular senescence. The Journal of Cell Biology, 193, 409–424. https://doi.org/10.1083/jcb.201010100.
Jazbutyte, V., Fiedler, J., Kneitz, S., Galuppo, P., Just, A., Holzmann, A., Bauersachs, J., & Thum, T. (2013). MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordrecht, Netherlands), 35, 747–762. https://doi.org/10.1007/s11357-012-9407-9.
Bar, N., & Dikstein, R. (2010). miR-22 forms a regulatory loop in PTEN/AKT pathway and modulates signaling kinetics. PloS One, 5, e10859. https://doi.org/10.1371/journal.pone.0010859.
Guo, M. M., Hu, L. H., Wang, Y. Q., Chen, P., Huang, J. G., Lu, N., He, J. H., & Liao, C. G. (2013). miR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1. Medical Oncology (Northwood, London, England), 30, 542. https://doi.org/10.1007/s12032-013-0542-7.
Kong, L. M., Liao, C. G., Zhang, Y., Xu, J., Li, Y., Huang, W., Zhang, Y., Bian, H., & Chen, Z. N. (2014). A regulatory loop involving miR-22, Sp1, and c-Myc modulates CD147 expression in breast cancer invasion and metastasis. Cancer Research, 74, 3764–3778. https://doi.org/10.1158/0008-5472.can-13-3555.
Song, S. J., Ito, K., Ala, U., Kats, L., Webster, K., Sun, S. M., Jongen-Lavrencic, M., Manova-Todorova, K., Teruya-Feldstein, J., Avigan, D. E., Delwel, R., & Pandolfi, P. P. (2013). The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell, 13, 87–101. https://doi.org/10.1016/j.stem.2013.06.003.
Chen, Z., Li, Q., Wang, S., & Zhang, J. (2015). miR4855p inhibits bladder cancer metastasis by targeting HMGA2. International Journal of Molecular Medicine, 36, 1136–1142. https://doi.org/10.3892/ijmm.2015.2302.
Wu, A., Wu, K., Li, J., Mo, Y., Lin, Y., Wang, Y., Shen, X., Li, S., Li, L., & Yang, Z. (2015). Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. Journal of Translational Medicine, 13, 105. https://doi.org/10.1186/s12967-015-0462-8.
Sampson, V. B., Rong, N. H., Han, J., Yang, Q., Aris, V., Soteropoulos, P., Petrelli, N. J., Dunn, S. P., & Krueger, L. J. (2007). MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Research, 67, 9762–9770. https://doi.org/10.1158/0008-5472.can-07-2462.
Chang, H. M., Martinez, N. J., Thornton, J. E., Hagan, J. P., Nguyen, K. D., & Gregory, R. I. (2012). Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nature Communications, 3, 923. https://doi.org/10.1038/ncomms1909.
Iwasaki, T., Tanaka, K., Kawano, M., Itonaga, I., & Tsumura, H. (2015). Tumor-suppressive microRNA-let-7a inhibits cell proliferation via targeting of E2F2 in osteosarcoma cells. International Journal of Oncology, 46, 1543–1550. https://doi.org/10.3892/ijo.2015.2867.
Faherty, N., Curran, S. P., O'Donovan, H., Martin, F., Godson, C., Brazil, D. P., & Crean, J. K. (2012). CCN2/CTGF increases expression of miR-302 microRNAs, which target the TGFbeta type II receptor with implications for nephropathic cell phenotypes. Journal of Cell Science, 125, 5621–5629. https://doi.org/10.1242/jcs.105528.
Keklikoglou, I., Koerner, C., Schmidt, C., Zhang, J. D., Heckmann, D., Shavinskaya, A., Allgayer, H., Guckel, B., Fehm, T., Schneeweiss, A., Sahin, O., Wiemann, S., & Tschulena, U. (2012). MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-kappaB and TGF-beta signaling pathways. Oncogene, 31, 4150–4163. https://doi.org/10.1038/onc.2011.571.
Subramanyam, D., Lamouille, S., Judson, R. L., Liu, J. Y., Bucay, N., Derynck, R., & Blelloch, R. (2011). Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology, 29, 443–448. https://doi.org/10.1038/nbt.1862.
Barroso-delJesus, A., Lucena-Aguilar, G., Sanchez, L., Ligero, G., Gutierrez-Aranda, I., & Menendez, P. (2011). The nodal inhibitor lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB Journal : Official Publication of the Federation of American Societies for Experimental Biology, 25, 1497–1508. https://doi.org/10.1096/fj.10-172221.
Rosa, A., Papaioannou, M. D., Krzyspiak, J. E., & Brivanlou, A. H. (2014). miR-373 is regulated by TGFbeta signaling and promotes mesendoderm differentiation in human embryonic stem cells. Developmental Biology, 391, 81–88. https://doi.org/10.1016/j.ydbio.2014.03.020.
Zhou, A. D., Diao, L. T., Xu, H., Xiao, Z. D., Li, J. H., Zhou, H., & Qu, L. H. (2012). Beta-catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/beta-catenin-signaling pathway. Oncogene, 31, 2968–2978. https://doi.org/10.1038/onc.2011.461.
Chen, Y. J., Luo, J., Yang, G. Y., Yang, K., Wen, S. Q., & Zou, S. Q. (2012). Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World Journal of Gastroenterology, 18, 3849–3861. https://doi.org/10.3748/wjg.v18.i29.3849.
Lee, M. R., Prasain, N., Chae, H. D., Kim, Y. J., Mantel, C., Yoder, M. C., & Broxmeyer, H. E. (2013). Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem cells (Dayton, Ohio), 31, 666–681. https://doi.org/10.1002/stem.1302.
Lee, K. H., Goan, Y. G., Hsiao, M., Lee, C. H., Jian, S. H., Lin, J. T., Chen, Y. L., & Lu, P. J. (2009). MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Experimental Cell Research, 315, 2529–2538. https://doi.org/10.1016/j.yexcr.2009.06.001.
Tian, Y., Liu, Y., Wang, T., Zhou, N., Kong, J., Chen, L., Snitow, M., Morley, M., Li, D., Petrenko, N., Zhou, S., Lu, M., Gao, E., Koch, W. J., Stewart, K. M., & Morrisey, E. E. (2015). A microRNA-hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Science Translational Medicine, 7, 279ra238. https://doi.org/10.1126/scitranslmed.3010841.
Acknowledgments
This research was supported by High Impact Research MOHE Grant UM.C/625/1/HIR/MOHE/DENT/01 from Ministry of Higher Education Malaysia, Fundamental Research Grant Scheme (FRGS FP044-2014B) from Ministry of Education, Malaysia and University of Malaya Research Grant (RP019C-13HTM) from University of Malaya. We would like to thank Prof. Dr. Sabri Musa for providing SHED samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have declared no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Aung, SW., Abu Kasim, N.H., Shamsuddin, S.A.A. et al. MicroRNAomic Transcriptomic Analysis Reveal Deregulation of Clustered Cellular Functions in Human Mesenchymal Stem Cells During in Vitro Passaging. Stem Cell Rev and Rep 16, 222–238 (2020). https://doi.org/10.1007/s12015-019-09924-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-019-09924-0