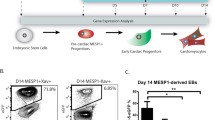
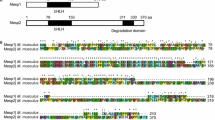
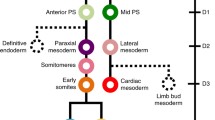
Abstract
The vertebrate early stage embryo is consisting of the three primary germ layers ectoderm, mesoderm and endoderm, from which all organ tissues are developed. During early embryonic development, mesodermal cells become sequentially determined to more precisely defined cell types including muscle, heart, vasculature, blood, kidney, gonads, dermis and cartilage. How the prospective mesodermal cells integrate the various signals they receive and how they resolve this information to regulate their morphogenetic behavior and cell fate decisions is largely unknown. Understanding of this complex phenomenon is essential to induce selective differentiation of pluripotent stem cells into clinically relevant, physiologically functional cells such as cardiomyocytes or for transdifferentiation of easily accessible cell types such as fibroblasts into other clinically relevant cell types for applications such as cell replacement therapy, accelerated drug discovery and drug toxicological testing. This demands an in-depth analysis of the mesodermal endogenous signaling cascades and transcription factor networks. Emerging results from isolation and transcriptome characterization of pure mesodermal cells derived from murine embryonic stem cells define the genetic and cellular identity of mesodermal cells and allows a comprehensive analysis of the very dynamic process of mesodermal patterning which would not be technically feasible with conventional embryology methods.
This review focuses on defining the transcriptomic signatures of mesodermal cells and their lineages with special reference to the molecular and signaling pathways associated with the complex process of mesodermal patterning.


Similar content being viewed by others
References
Doss, M. X., Wagh, V., Schulz, H., Kull, M., Kolde, R., Pfannkuche, K., et al. (2010). Global transcriptomic analysis of murine embryonic stem cell-derived brachyury (T) cells. Genes to Cells, 15, 209–228.
Doss, M. X., Chen, S., Winkler, J., Hippler-Altenburg, R., Odenthal, M., Wickenhauser, C., et al. (2007). Transcriptomic and phenotypic analysis of murine embryonic stem cell derived BMP2+ lineage cells: an insight into mesodermal patterning. Genome Biology, 8(9), R184.
Doss, M. X., Winkler, J., Chen, S., Hippler-Altenburg, R., Sotiriadou, I., Halbach, M., et al. (2007). Global transcriptome analysis of murine embryonic stem cell-derived cardiomyocytes. Genome Biology, 8(4), R56.
Willems, E., & Leyns, L. (2008). Patterning of mouse embryonic stem cell-derived pan-mesoderm by Activin A/Nodal and Bmp4 signaling requires Fibroblast Growth Factor activity. Differentiation, 76(7), 745–759.
Watson, C. M., & Tam, P. P. (2001). Cell lineage determination in the mouse. Cell Structure and Function, 26(3), 123–129.
Parameswaran, M., & Tam, P. P. (1995). Regionalisation of cell fate and morphogenetic movement of the mesoderm during mouse gastrulation. Developmental Genetics, 17(1), 16–28.
Burdsal, C. A., Damsky, C. H., & Pedersen, R. A. (1993). The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development, 118(3), 829–844.
Poelmann, R. E. (1981). The formation of the embryonic mesoderm in the early post-implantation mouse embryo. Anat Embryol (Berl), 162(1), 29–40.
Tam, P. P., & Behringer, R. R. (1997). Mouse gastrulation: the formation of a mammalian body plan. Mechanisms of Development, 68(1–2), 3–25.
Kispert, A., & Herrmann, B. G. (1994). Immunohistochemical analysis of the Brachyury protein in wild-type and mutant mouse embryos. Developmental Biology, 161(1), 179–193.
Herrmann, B. G. (1991). Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development, 113(3), 913–917.
Fehling, H. J., Lacaud, G., Kubo, A., Kennedy, M., Robertson, S., Keller, G., et al. (2003). Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development, 130(17), 4217–4227.
Kinder, S. J., Tsang, T. E., Quinlan, G. A., Hadjantonakis, A. K., Nagy, A., & Tam, P. P. (1999). The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development, 126(21), 4691–4701.
Gadue, P., Huber, T. L., Nostro, M. C., Kattman, S., & Keller, G. M. (2005). Germ layer induction from embryonic stem cells. Experimental Hematology, 33(9), 955–964.
Doss, M. X., Koehler, C. I., Gissel, C., Hescheler, J., & Sachinidis, A. (2004). Embryonic stem cells: a promising tool for cell replacement therapy. Journal of Cellular and Molecular Medicine, 8(4), 465–473.
de Peppo, G. M., Svensson, S., Lenneras, M., Synnergren, J., Stenberg, J., Strehl, R., et al. (2010). Human embryonic mesodermal progenitors highly resemble human mesenchymal stem cells and display high potential for tissue engineering applications. Tissue Engineering. Part A, 16(7), 2161–2182.
Gunaseeli, I., Doss, M. X., Antzelevitch, C., Hescheler, J., & Sachinidis, A. (2010). Induced pluripotent stem cells as a model for accelerated patient- and disease-specific drug discovery. Current Medicinal Chemistry, 17(8), 759–766.
Potta, S. P., Liang, H., Winkler, J., Doss, M. X., Chen, S., Wagh, V., et al. (2010). Isolation and functional characterization of alpha-smooth muscle actin expressing cardiomyocytes from embryonic stem cells. Cellular Physiology and Biochemistry, 25(6), 595–604.
Mariappan, D., Winkler, J., Chen, S., Schulz, H., Hescheler, J., & Sachinidis, A. (2009). Transcriptional profiling of CD31(+) cells isolated from murine embryonic stem cells. Genes to Cells, 14(2), 243–260.
Weinhold, B., Schratt, G., Arsenian, S., Berger, J., Kamino, K., Schwarz, H., et al. (2000). Srf(-/-) ES cells display non-cell-autonomous impairment in mesodermal differentiation. The EMBO Journal, 19(21), 5835–5844.
Zhang, H., & Bradley, A. (1996). Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development, 122(10), 2977–2986.
Ghatpande, S., Brand, T., Zile, M., & Evans, T. (2006). Bmp2 and Gata4 function additively to rescue heart tube development in the absence of retinoids. Developmental Dynamics, 235(8), 2030–2039.
Schultheiss, T. M., & Lassar, A. B. (1997). Induction of chick cardiac myogenesis by bone morphogenetic proteins. Cold Spring Harbor Symposia on Quantitative Biology, 62, 413–419.
Kimura, N., Matsuo, R., Shibuya, H., Nakashima, K., & Taga, T. (2000). BMP2-induced apoptosis is mediated by activation of the TAK1-p38 kinase pathway that is negatively regulated by Smad6. The Journal of Biological Chemistry, 275(23), 17647–17652.
Hattori, A., Katayama, M., Iwasaki, S., Ishii, K., Tsujimoto, M., & Kohno, M. (1999). Bone morphogenetic protein-2 promotes survival and differentiation of striatal GABAergic neurons in the absence of glial cell proliferation. Journal of Neurochemistry, 72(6), 2264–2271.
Iwasaki, S., Hattori, A., Sato, M., Tsujimoto, M., & Kohno, M. (1996). Characterization of the bone morphogenetic protein-2 as a neurotrophic factor. Induction of neuronal differentiation of PC12 cells in the absence of mitogen-activated protein kinase activation. The Journal of Biological Chemistry, 271(29), 17360–17365.
Potta, S. P., Liang, H., Pfannkuche, K., Winkler, J., Chen, S., Doss, M. X., et al. (2009). Functional characterization and transcriptome analysis of embryonic stem cell-derived contractile smooth muscle cells. Hypertension, 53(2), 196–204.
Moretti, A., Caron, L., Nakano, A., Lam, J. T., Bernshausen, A., Chen, Y., et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell, 127(6), 1151–1165.
Yang, L., Soonpaa, M. H., Adler, E. D., Roepke, T. K., Kattman, S. J., Kennedy, M., et al. (2008). Human cardiovascular progenitor cells develop from a KDR + embryonic-stem-cell-derived population. Nature, 453(7194), 524–528.
Yoshikawa, Y., Fujimori, T., McMahon, A. P., & Takada, S. (1997). Evidence that absence of Wnt-3a signaling promotes neuralization instead of paraxial mesoderm development in the mouse. Developmental Biology, 183(2), 234–242.
Loebel, D. A., Watson, C. M., De Young, R. A., & Tam, P. P. (2003). Lineage choice and differentiation in mouse embryos and embryonic stem cells. Developmental Biology, 264(1), 1–14.
Galceran, J., Farinas, I., Depew, M. J., Clevers, H., & Grosschedl, R. (1999). Wnt3a-/–like phenotype and limb deficiency in Lef1(-/-)Tcf1(-/-) mice. Genes & Development, 13(6), 709–717.
Yamaguchi, T. P., Bradley, A., McMahon, A. P., & Jones, S. (1999). A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development, 126(6), 1211–1223.
Galceran, J., Hsu, S. C., & Grosschedl, R. (2001). Rescue of a Wnt mutation by an activated form of LEF-1: regulation of maintenance but not initiation of Brachyury expression. Proceedings of the National Academy of Sciences of the United States of America, 98(15), 8668–8673.
Ciruna, B., & Rossant, J. (2001). FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Developmental Cell, 1(1), 37–49.
Chapman, D. L., & Papaioannou, V. E. (1998). Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature, 391(6668), 695–697.
Ciruna, B. G., Schwartz, L., Harpal, K., Yamaguchi, T. P., & Rossant, J. (1997). Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development, 124(14), 2829–2841.
Sun, X., Meyers, E. N., Lewandoski, M., & Martin, G. R. (1999). Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes & Development, 13(14), 1834–1846.
Yamaguchi, T. P., Harpal, K., Henkemeyer, M., & Rossant, J. (1994). fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Development, 8(24), 3032–3044.
Winnier, G., Blessing, M., Labosky, P. A., & Hogan, B. L. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & Development, 9(17), 2105–2116.
Mishina, Y., Suzuki, A., Ueno, N., & Behringer, R. R. (1995). Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes & Development, 9(24), 3027–3037.
Wilson, V., Rashbass, P., & Beddington, R. S. (1993). Chimeric analysis of T (Brachyury) gene function. Development, 117(4), 1321–1331.
Miyazawa, K., Shinozaki, M., Hara, T., Furuya, T., & Miyazono, K. (2002). Two major Smad pathways in TGF-beta superfamily signalling. Genes to Cells, 7(12), 1191–1204.
Yang, Y., Guillot, P., Boyd, Y., Lyon, M. F., & McMahon, A. P. (1998). Evidence that preaxial polydactyly in the Doublefoot mutant is due to ectopic Indian Hedgehog signaling. Development, 125(16), 3123–3132.
Sirard, C., de la Pompa, J. L., Elia, A., Itie, A., Mirtsos, C., Cheung, A., et al. (1998). The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes & Development, 12(1), 107–119.
Bachiller, D., Klingensmith, J., Kemp, C., Belo, J. A., Anderson, R. M., May, S. R., et al. (2000). The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature, 403(6770), 658–661.
Locascio, A., & Nieto, M. A. (2001). Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Current Opinion in Genetics & Development, 11(4), 464–469.
Ding, J., Yang, L., Yan, Y. T., Chen, A., Desai, N., Wynshaw-Boris, A., et al. (1998). Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature, 395(6703), 702–707.
Meno, C., Gritsman, K., Ohishi, S., Ohfuji, Y., Heckscher, E., Mochida, K., et al. (1999). Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Molecular Cell, 4(3), 287–298.
Johnson, W. E., Li, C., & Rabinovic, A. (2007). Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1), 118–127.
Adler, P., Reimand, J., Janes, J., Kolde, R., Peterson, H., & Vilo, J. (2008). KEGGanim: pathway animations for high-throughput data. Bioinformatics, 24(4), 588–590.
Acknowledgement
This work was supported by a grant from the European Community (6th Framework Programme, Thematic Priority: Life sciences, genomics and biotechnology for health; contract no: FunGenES LSHG-CT-2003-503494).
Disclosure
The authors declare no potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Below is the link to the electronic supplementary material.
Supplementary Table 1
Enriched Gene ontology annotations in k means clusters (XLS 289 kb)
Supplementary Figure 1
KEGGanim tool is used to color the pathway members (genes) at these three KEGG pathways; Wnt signaling Pathway, TGFβ signaling pathway and Hedgehog signaling pathways. The genes are colored red and green according to their expression signal values as found at the dataset having all the five pure populations such as αMHC+ cardiomyocytes, BMP2+ and Brachyury+ mesodermal cells, CD31+ endothelial-like and Acta2+ smooth cells. The red color indicates the high expression signal and green the low expression signal. If several probesets or proteins match a pathway member, the corresponding node is split into smaller colored areas to reflect different experimental values [51]. (PPT 585 kb)
Rights and permissions
About this article
Cite this article
Doss, M.X., Gaspar, J.A., Winkler, J. et al. Specific Gene Signatures and Pathways in Mesodermal Cells and Their Derivatives Derived from Embryonic Stem Cells. Stem Cell Rev and Rep 8, 43–54 (2012). https://doi.org/10.1007/s12015-011-9263-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12015-011-9263-5