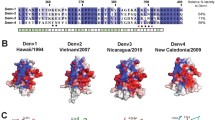
Abstract
The dengue virus (DENV), composed of four distinct but serologically related Flaviviruses, causes the most important emerging viral disease, with nearly 400 million infections yearly. Currently, there are no approved therapies. Although DENV infection induces lifelong immunity against the same serotype, the antibodies raised contribute to severe disease in heterotypic infections. Therefore, understanding the mechanism of DENV neutralization by antibodies is crucial in the design of vaccines against all serotypes. This study reports a comparative structural and energetic analysis of the monoclonal antibody (mAb) 4E11 in complex with its target domain III of the envelope protein for all four DENV serotypes. We use extensive replica molecular dynamics simulations in conjunction with the binding free energy calculations. Further single point and double mutations were designed through computational site-directed mutagenesis and observed that the re-engineered antibody exhibits high affinity to binding and broadly neutralizing activity against serotypes. Our results showed improved binding affinity by the gain of enthalpy, which could be attributed to the stabilization of salt-bridge and hydrogen bond interactions at the antigen-antibody interface. The findings provide valuable results in understanding the structural dynamics and energetic contributions that will be helpful to the design of high-affinity antibodies against dengue infections.










Similar content being viewed by others
Data availability
The data supporting this study’s findings are available from the corresponding author (PK) upon reasonable request.
References
Monath, T. P. (1994). Dengue: the risk to developed and developing countries. Proceedings of the National Academy of Sciences, 91(7), 2395–2400. https://doi.org/10.1073/pnas.91.7.2395.
Bhatt, S., Gething, P. W., Brady, O. J., Messina, J. P., Farlow, A. W., Moyes, C. L., & Hay, S. I. (2013). The global distribution and burden of dengue. Nature, 496(7446), 504–507. https://doi.org/10.1038/nature12060.
Shepard, D. S., Undurraga, E. A., Halasa, Y. A., & Stanaway, J. D. (2016). The global economic burden of dengue: a systematic analysis. The Lancet Infectious Diseases, 16(8), 935–941. https://doi.org/10.1016/S1473-3099(16)00146-8.
Mackenzie, J. S., Gubler, D. J., & Petersen, L. R. (2004). Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nature Medicine, 10(S12), S98–S109. https://doi.org/10.1038/nm1144.
Sabin, A. B. (1952). Research on Dengue during World War II 1. The American Journal of Tropical Medicine and Hygiene, 1(1), 30–50. https://doi.org/10.4269/ajtmh.1952.1.30.
Sangkawibha, N., Rojanasuphot, S., Ahandrik, S., Viriyapongse, S., Jatanasen, S., Salitul, V., & Halstead, S. B. (1984). RISK FACTORS IN DENGUE SHOCK SYNDROME: A PROSPECTIVE EPIDEMIOLOGIC STUDY IN RAYONG, THAILAND. American Journal of Epidemiology, 120(5), 653–669. https://doi.org/10.1093/oxfordjournals.aje.a113932.
Halstead, S., & O’Rourke, E. (1977). Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. Journal of Experimental Medicine, 146(1), 201–217. https://doi.org/10.1084/jem.146.1.201.
Capeding, M. R., Tran, N. H., Hadinegoro, S. R. S., Ismail, H. I. H. M., Chotpitayasunondh, T., Chua, M. N., & Bouckenooghe, A. (2014). Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. The Lancet, 384(9951), 1358–1365. https://doi.org/10.1016/S0140-6736(14)61060-6.
Sabchareon, A., Wallace, D., Sirivichayakul, C., Limkittikul, K., Chanthavanich, P., Suvannadabba, S., & Lang, J. (2012). Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. The Lancet, 380(9853), 1559–1567. https://doi.org/10.1016/S0140-6736(12)61428-7.
Villar, L., Dayan, G. H., Arredondo-García, J. L., Rivera, D. M., Cunha, R., Deseda, C., & Noriega, F. (2015). Efficacy of a Tetravalent Dengue Vaccine in Children in Latin America. New England Journal of Medicine, 372(2), 113–123. https://doi.org/10.1056/NEJMoa1411037.
Perera, R., & Kuhn, R. J. (2008). Structural proteomics of dengue virus. Current Opinion in Microbiology, 11(4), 369–377. https://doi.org/10.1016/j.mib.2008.06.004.
Crill, W. D., & Roehrig, J. T. (2001). Monoclonal Antibodies That Bind to Domain III of Dengue Virus E Glycoprotein Are the Most Efficient Blockers of Virus Adsorption to Vero Cells. Journal of Virology, 75(16), 7769–7773. https://doi.org/10.1128/JVI.75.16.7769-7773.2001.
Caskey, M., Klein, F., Lorenzi, J. C. C., Seaman, M. S., West, A. P., Buckley, N., & Nussenzweig, M. C. (2015). Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature, 522(7557), 487–491. https://doi.org/10.1038/nature14411.
Ramos, E. L., Mitcham, J. L., Koller, T. D., Bonavia, A., Usner, D. W., Balaratnam, G., & Swiderek, K. M. (2015). Efficacy and Safety of Treatment With an Anti-M2e Monoclonal Antibody in Experimental Human Influenza. Journal of Infectious Diseases, 211(7), 1038–1044. https://doi.org/10.1093/infdis/jiu539.
Siber, G. R., Werner, B. G., Halsey, N. A., Reid, R., Almeido-Hill, J., Garrett, S. C., & Santosham, M. (1993). Interference of immune globulin with measles and rubella immunization. The Journal of Pediatrics, 122(2), 204–211. https://doi.org/10.1016/S0022-3476(06)80114-9.
Siegrist, C.-A., Córdova, M., Brandt, C., Barrios, C., Berney, M., Tougne, C., & Lambert, P.-H. (1998). Determinants of infant responses to vaccines in presence of maternal antibodies. Vaccine, 16(14–15), 1409–1414. https://doi.org/10.1016/S0264-410X(98)00100-5.
Smith, S. A., de Alwis, A. R., Kose, N., Harris, E., Ibarra, K. D., Kahle, K. M., & Crowe, J. E. (2013). The Potent and Broadly Neutralizing Human Dengue Virus-Specific Monoclonal Antibody 1C19 Reveals a Unique Cross-Reactive Epitope on the bc Loop of Domain II of the Envelope Protein. mBio, 4(6), e00873–13. https://doi.org/10.1128/mBio.00873-13.
Lai, C.-Y., Tsai, W.-Y., Lin, S.-R., Kao, C.-L., Hu, H.-P., King, C.-C., & Wang, W.-K. (2008). Antibodies to Envelope Glycoprotein of Dengue Virus during the Natural Course of Infection Are Predominantly Cross-Reactive and Recognize Epitopes Containing Highly Conserved Residues at the Fusion Loop of Domain II. Journal of Virology, 82(13), 6631–6643. https://doi.org/10.1128/JVI.00316-08.
Beltramello, M., Williams, K. L., Simmons, C. P., Macagno, A., Simonelli, L., Quyen, N. T. H., & Sallusto, F. (2010). The Human Immune Response to Dengue Virus Is Dominated by Highly Cross-Reactive Antibodies Endowed with Neutralizing and Enhancing Activity. Cell Host & Microbe, 8(3), 271–283. https://doi.org/10.1016/j.chom.2010.08.007.
Dejnirattisai, W., Wongwiwat, W., Supasa, S., Zhang, X., Dai, X., Rouvinski, A., & Screaton, G. R. (2015). A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nature Immunology, 16(2), 170–177. https://doi.org/10.1038/ni.3058.
de Alwis, R., Smith, S. A., Olivarez, N. P., Messer, W. B., Huynh, J. P., Wahala, W. M. P. B., & de Silva, A. M. (2012). Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proceedings of the National Academy of Sciences, 109(19), 7439–7444. https://doi.org/10.1073/pnas.1200566109.
Wahala, W. M. P. B., Kraus, A. A., Haymore, L. B., Accavitti-Loper, M. A., & de Silva, A. M. (2009). Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology, 392(1), 103–113. https://doi.org/10.1016/j.virol.2009.06.037.
Williams, K. L., Wahala, W. M. P. B., Orozco, S., de Silva, A. M., & Harris, E. (2012). Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology, 429(1), 12–20. https://doi.org/10.1016/j.virol.2012.03.003.
Sukupolvi-Petty, S., Austin, S. K., Engle, M., Brien, J. D., Dowd, K. A., Williams, K. L., & Diamond, M. S. (2010). Structure and Function Analysis of Therapeutic Monoclonal Antibodies against Dengue Virus Type 2. Journal of Virology, 84(18), 9227–9239. https://doi.org/10.1128/JVI.01087-10.
Sukupolvi-Petty, S., Austin, S. K., Purtha, W. E., Oliphant, T., Nybakken, G. E., Schlesinger, J. J., & Diamond, M. S. (2007). Type- and Subcomplex-Specific Neutralizing Antibodies against Domain III of Dengue Virus Type 2 Envelope Protein Recognize Adjacent Epitopes. Journal of Virology, 81(23), 12816–12826. https://doi.org/10.1128/JVI.00432-07.
Sabin, A. B., & Schlesinger, R. W. (1945). Production of Immunity to Dengue with Virus Modified by Propagation in Mice. Science, 101(2634), 640–642. https://doi.org/10.1126/science.101.2634.640.
Hadinegoro, S. R., Arredondo-García, J. L., Capeding, M. R., Deseda, C., Chotpitayasunondh, T., Dietze, R., & Saville, M. (2015). Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. New England Journal of Medicine, 373(13), 1195–1206. https://doi.org/10.1056/NEJMoa1506223.
Kirkpatrick, B. D., Durbin, A. P., Pierce, K. K., Carmolli, M. P., Tibery, C. M., Grier, P. L., & Whitehead, S. S. (2015). Robust and Balanced Immune Responses to All 4 Dengue Virus Serotypes Following Administration of a Single Dose of a Live Attenuated Tetravalent Dengue Vaccine to Healthy, Flavivirus-Naive Adults. Journal of Infectious Diseases, 212(5), 702–710. https://doi.org/10.1093/infdis/jiv082.
Dejnirattisai, W., Jumnainsong, A., Onsirisakul, N., Fitton, P., Vasanawathana, S., Limpitikul, W., & Screaton, G. (2010). Cross-Reacting Antibodies Enhance Dengue Virus Infection in Humans. Science, 328(5979), 745–748. https://doi.org/10.1126/science.1185181.
Thullier, P., Lafaye, P., Mégret, F., Deubel, V., Jouan, A., & Mazié, J. C. (1999). A recombinant Fab neutralizes dengue virus in vitro. Journal of Biotechnology, 69(2–3), 183–190. https://doi.org/10.1016/S0168-1656(99)00037-1.
Thullier, P., Demangel, C., Bedouelle, H., Mégret, F., Jouan, A., Deubel, V., & Lafaye, P. (2001). Mapping of a dengue virus neutralizing epitope critical for the infectivity of all serotypes: insight into the neutralization mechanism. Journal of General Virology, 82(8), 1885–1892. https://doi.org/10.1099/0022-1317-82-8-1885.
Cockburn, J. J. B., Navarro Sanchez, M. E., Fretes, N., Urvoas, A., Staropoli, I., Kikuti, C. M., & Rey, F. A. (2012). Mechanism of Dengue Virus Broad Cross-Neutralization by a Monoclonal Antibody. Structure, 20(2), 303–314. https://doi.org/10.1016/j.str.2012.01.001.
Gromowski, G. D., Roehrig, J. T., Diamond, M. S., Lee, J. C., Pitcher, T. J., & Barrett, A. D. T. (2010). Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology, 407(2), 237–246. https://doi.org/10.1016/j.virol.2010.06.044.
Matsui, K., Gromowski, G. D., Li, L., & Barrett, A. D. T. (2010). Characterization of a dengue type-specific epitope on dengue 3 virus envelope protein domain III. Journal of General Virology, 91(9), 2249–2253. https://doi.org/10.1099/vir.0.021220-0.
Shrestha, B., Brien, J. D., Sukupolvi-Petty, S., Austin, S. K., Edeling, M. A., Kim, T., & Diamond, M. S. (2010). The Development of Therapeutic Antibodies That Neutralize Homologous and Heterologous Genotypes of Dengue Virus Type 1. PLoS Pathogens, 6(4), e1000823 https://doi.org/10.1371/journal.ppat.1000823.
Rey, F. A., Heinz, F. X., Mandl, C., Kunz, C., & Harrison, S. C. (1995). The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature, 375(6529), 291–298. https://doi.org/10.1038/375291a0.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera?A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/10.1002/jcc.20084.
Case, D. A., Ben-Shalom, I. Y., Brozell, S. R., Cerutti, D. S., Cheatham, III, T. E., Cruzeiro, V. W. D., … Kollman, P. A. (2018). AMBER 2018. University of California, San Francisco.
Price, D. J., & Brooks, C. L. (2004). A modified TIP3P water potential for simulation with Ewald summation. The Journal of Chemical Physics, 121(20), 10096–10103. https://doi.org/10.1063/1.1808117.
Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. https://doi.org/10.1021/acs.jctc.5b00255.
Kräutler, V., Gunsteren, W. Fvan, & Hünenberger, P. H. (2001). A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of Computational Chemistry, 22(5), 501–508. https://doi.org/10.1002/1096-987X(20010415)22:5<501::AID-JCC1021>3.0.CO;2-V
Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N ⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. https://doi.org/10.1063/1.464397.
Loncharich, R. J., Brooks, B. R., & Pastor, R. W. (1992). Langevin dynamics of peptides: The frictional dependence of isomerization rates ofN-acetylalanyl-N?-methylamide. Biopolymers, 32(5), 523–535. https://doi.org/10.1002/bip.360320508.
Pastor, R. W., Brooks, B. R., & Szabo, A. (1988). An analysis of the accuracy of Langevin and molecular dynamics algorithms. Molecular Physics, 65(6), 1409–1419. https://doi.org/10.1080/00268978800101881.
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. https://doi.org/10.1063/1.448118.
Roe, D. R., & Cheatham, T. E. (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. https://doi.org/10.1021/ct400341p.
Dannenberg, J. J. (1998). An Introduction to Hydrogen Bonding By George A. Jeffrey (University of Pittsburgh). Oxford University Press: New York and Oxford. 1997. ix + 303 pp. $60.00. ISBN 0-19-509549-9. Journal of the American Chemical Society, 120(22), 5604–5604. https://doi.org/10.1021/ja9756331.
Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., & Cheatham, T. E. (2000). Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Accounts of Chemical Research, 33(12), 889–897. https://doi.org/10.1021/ar000033j.
Miller, B. R., McGee, T. D., Swails, J. M., Homeyer, N., Gohlke, H., & Roitberg, A. E. (2012). MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. Journal of Chemical Theory and Computation, 8(9), 3314–3321. https://doi.org/10.1021/ct300418h.
Hou, T., Wang, J., Li, Y., & Wang, W. (2011). Assessing the Performance of the MM/PBSA and MM/GBSA Methods. 1. The Accuracy of Binding Free Energy Calculations Based on Molecular Dynamics Simulations. Journal of Chemical Information and Modeling, 51(1), 69–82. https://doi.org/10.1021/ci100275a.
Jonniya, N. A., Sk, M. F., & Kar, P. (2020). A comparative study of structural and conformational properties of WNK kinase isoforms bound to an inhibitor: insights from molecular dynamic simulations. Journal of Biomolecular Structure and Dynamics, 0(0), 1–16. https://doi.org/10.1080/07391102.2020.1827035.
Kar, P., Lipowsky, R., & Knecht, V. (2013). Importance of Polar Solvation and Configurational Entropy for Design of Antiretroviral Drugs Targeting HIV-1 Protease. The Journal of Physical Chemistry B, 117(19), 5793–5805. https://doi.org/10.1021/jp3085292.
Amarnath Jonniya, N., Fulbabu Sk, M., & Kar, P. (2021). Characterizing an allosteric inhibitor-induced inactive state in with-no-lysine kinase 1 using Gaussian accelerated molecular dynamics simulations. Physical Chemistry Chemical Physics, 23(12), 7343–7358. https://doi.org/10.1039/D0CP05733A.
Jonniya, N. A., Sk, M. F., & Kar, P. (2021). Elucidating specificity of an allosteric inhibitor WNK476 among With-No-Lysine kinase isoforms using molecular dynamic simulations. Chemical Biology & Drug Design, 98(3), 405–420. https://doi.org/10.1111/cbdd.13863.
Jonniya, N. A., Sk, M. F., & Kar, P. (2019). Investigating Phosphorylation-Induced Conformational Changes in WNK1 Kinase by Molecular Dynamics Simulations. ACS Omega, 4(17), 17404–17416. https://doi.org/10.1021/acsomega.9b02187.
Jonniya, N. A., & Kar, P. (2022). Functional Loop Dynamics and Characterization of the Inactive State of the NS2B-NS3 Dengue Protease due to Allosteric Inhibitor Binding. Journal of Chemical Information and Modeling, 62(16), 3800–3813. https://doi.org/10.1021/acs.jcim.2c00461.
Roy, R., Jonniya, N. A., Poddar, S., Sk, M. F., & Kar, P. (2021). Unraveling the Molecular Mechanism of Recognition of Human Interferon-Stimulated Gene Product 15 by Coronavirus Papain-Like Proteases: A Multiscale Simulation Study. Journal of Chemical Information and Modeling. https://doi.org/10.1021/acs.jcim.1c00918
Lee, V. S., Tue-ngeun, P., Nangola, S., Kitidee, K., Jitonnom, J., Nimmanpipug, P., & Tayapiwatana, C. (2010). Pairwise decomposition of residue interaction energies of single chain Fv with HIV-1 p17 epitope variants. Molecular Immunology, 47(5), 982–990. https://doi.org/10.1016/j.molimm.2009.11.021.
Ieong, P., Amaro, R. E., & Li, W. W. (2015). Molecular Dynamics Analysis of Antibody Recognition and Escape by Human H1N1 Influenza Hemagglutinin. Biophysical Journal, 108(11), 2704–2712. https://doi.org/10.1016/j.bpj.2015.04.025.
Gohlke, H., Kiel, C., & Case, D. A. (2003). Insights into Protein–Protein Binding by Binding Free Energy Calculation and Free Energy Decomposition for the Ras–Raf and Ras–RalGDS Complexes. Journal of Molecular Biology, 330(4), 891–913. https://doi.org/10.1016/S0022-2836(03)00610-7.
Rodrigues, C. H., Pires, D. E. & & Ascher, D. B. (2018). DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Research, 46(W1), W350–W355. https://doi.org/10.1093/nar/gky300.
Pires, D. E. V., Ascher, D. B. & & Blundell, T. L. (2014). DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Research, 42(W1), W314–W319. https://doi.org/10.1093/nar/gku411.
da Silveira, C. H., Pires, D. E. V., Minardi, R. C., Ribeiro, C., Veloso, C. J. M., Lopes, J. C. D., & Santoro, M. M. (2009). Protein cut-off scanning: A comparative analysis of cut-off dependent and cut-off free methods for prospecting contacts in proteins. Proteins: Structure, Function, and Bioinformatics, 74(3), 727–743. https://doi.org/10.1002/prot.22187.
Pires, D. E. V., Ascher, D. B., & Blundell, T. L. (2014). mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics, 30(3), 335–342. https://doi.org/10.1093/bioinformatics/btt691.
Acknowledgements
This work was supported by the Department of Science and Technology, Govt. of India (grant number DST/NSM/R&D_HPC_Applications/2021/03.18).
Author contributions
N.A.J. and P.K.: Conceptualization; N.A.J.: Writing- Original Draft, Methodology, Visualization, Formal analysis, Software, Validation, Investigation; N.A.J. and P.K.: Writing-Review and Editing, Validation; P.K.: Methodology, Software, Resources, Funding acquisition, Project administration. All authors have read and agreed to the published version of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jonniya, N.A., Poddar, S., Mahapatra, S. et al. Computer-aided Affinity Enhancement of a Cross-reactive Antibody against Dengue Virus Envelope Domain III. Cell Biochem Biophys 81, 737–755 (2023). https://doi.org/10.1007/s12013-023-01175-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-023-01175-8