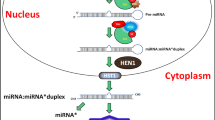
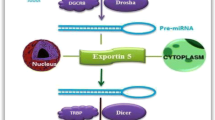
Abstract
Anthropogenic activities have contributed hugely in enhancing various types of environmental toxicity. One of these is higher accumulation of toxic heavy metals in soil and plant tissues. Although many heavy metals act as essential component for the growth and development of plants when present in low concentrations but at higher concentrations it becomes cytotoxic. Several innate mechanisms have evolved in plants to cope with it. In recent years the mechanism of using miRNA to combat metal induced toxicity has come to fore front. The miRNA or the microRNA regulates different physiological processes and induces a negative control in expressing the complementary target genes. The cleavage formation by post-transcriptional method and the inhibition of targeted translational mRNA are the two main procedures by which plant miRNAs function. The heavy and enhanced metal accumulation in plants has increased the production of different kinds of free radicals like reactive nitrogen and oxygen which damage the plants oxidatively. Several plant miRNA are capable of targeting and reducing the expression of those genes which are responsible for higher metal accumulation and storage. This can reduce the metal load and hence its negative impact on plant can also be reduced. This review depicts the biogenesis, the mode of action of miRNA, and the control mechanisms of miRNA in metal induced stress response in plant. A detailed review on the role of plant miRNA in alleviation of metal induced stress is discussed in this present study.










Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Sanz-Carbonell, A., Marques, M. C., Bustamante, A., Fares, M. A., Rodrigo, G., & Gomez, G. (2019). Inferring the regulatory network of the miRNA-mediated response to biotic and abiotic stress in melon. BMC Plant Biology, 19, 78.
Srivastava, S., & Suprasanna, P. (2021). MicroRNAs: Tiny, powerful players of metal stress responses in plants. Plant Physiology and Biochemistry, 166, 928–938.
DalCorso, G., Farinati, S., & Furini, S. (2010). Regulatory networks of cadmium stress in plants. Plant Signaling & Behavior, 5(6), 663–667.
Patra, M., Bhowmik, N., Bandopadhyay, B., & Sharma, A. (2004). Comparison of mercury, lead and arsenic with respect to genotoxic effects on plant systems and the development of genetic tolerance. Environmental and Experimental Botany, 52(3), 199–223.
Yruela, I. (2009). Copper in plants: Acquisition, transport and interactions. Functional Plant Biology, 36(5), 409–430.
Hall, J. L. (2011). Cellular mechanisms for heavy metal detoxification and tolerance. Journal of Experimental Botany, 53, 1–11.
Seth, C. S., Remans, T., Keunen, E., Jozefczak, M., Gielen, H., Opdenakker, K., Weyens, N., Vangronsveld, J., & Cuypers, A. (2012). Phytoextraction of toxic metals: A central role for glutathione. Plant, Cell & Environment, 35, 334–346.
Smeets, K., Opdenakker, K., Remans, T., van Sanden, S., van Belleghem, F., Semane, B., Horemans, N., Guisez, Y., Vangronsveld, J., & Cuypers, A. (2009). Oxidative stress-related responses at transcriptional and enzymatic levels after exposure to cd or Cu in a multipollution context. Journal of Plant Physiology, 166, 1982–1992.
Cuypers, A., Smeets, K., Ruytinx, J., Opdenakker, K., Keunen, E., Remans, T., Horemans, N., Vanhoudt, N., van Sanden, S., van Belleghem, F., et al. (2011). The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. Journal of Plant Physiology, 168, 309–316.
Shinozaki, K., & Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany, 58(2), 221–227.
Drazkiewicz, M., Skórzynska-Polit, E., & Krupa, Z. (2011). Effect of BSO-supplemented heavy metals on antioxidant enzymes in Arabidopsis thaliana. Ecotoxicology and Environmental Safety, 73, 1362–1369.
Liu, Y., Teng, C., Xia, R., & Meyers, B. C. (2020). PhasiRNAs in plants: Their biogenesis, genic sources, and roles in stress responses, development, and reproduction. The Plant Cell, 32, 3059–3080.
Khraiwesh, B., Zhu, J. K., & Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica Et Biophysica Acta, 1819(2), 137–148.
Jamalkandi, S. A., & Masoudi-Nejad, A. (2009). Reconstruction of Arabidopsis thaliana fully integrated small RNA pathway. Functional and Integrative Genomics, 9, 419–432.
Sunkar, R., Kapoor, A., & Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by down regulation of miR398 and important for oxidative stress tolerance. The Plant Cell, 18(8), 2051–2065.
Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H., & Poethig, R. S. (2012). Nuclear processing and export of microRNAs in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 102, 3691–3696.
Vaucheret, H., Vazquez, F., Crété, P., & Bartel, D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Development, 18, 1187–1197.
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., & Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell, 110(4), 513–20.
Mallory-Smith, C., & Zapiola, M. (2008). Gene flow from glyphosate-resistant crops. Pest Management Science, 64(4), 428–440.
Vazquez, F., Blevins, T., Ailhas, J., Boller, T., & Meins, F., Jr. (2008). Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Research, 36, 6429–6438.
Chellappan, P., Xia, J., Zhou, X., Gao, S., Zhang, X., Coutino, G., Vazquez, F., Zhang, W., & Jin, H. (2010). siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Research, 38, 6883–6894.
Matzke, M., Kanno, T., Daxinger, L., Huettel, B., & Matzke, A. J. (2009). RNA-mediated chromatin-based silencing in plants. Current Opinion in Cell Biology, 21, 367–376. https://doi.org/10.1016/j.ceb.2009.01.025
Bao, N., Lye, K. W., & Barton, M. K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Developmental Cell, 7, 653–662.
Boyko, A., & Kovalchuk, I. (2008). Epigenetic control of plant stress response. Environmental and Molecular Mutagenesis, 49, 61–72.
Luo, M., Liu, X., Singh, P., Cui, Y., Zimmerli, L., & Wu, K. (2012). Chromatin modifications and remodeling in plant abiotic stress responses. Biochimica et Biophysica Acta, 1819, 129–136.
Cao, D., Gao, X., Liu, J., Wang, X., Geng, S., Yang, C., Liu, B., & Shi, D. (2012). Root-specific DNA methylation in Chloris virgata, a natural alkaline-resistant halophyte, in response to salt and alkaline stresses. Plant Molecular Biology Reporter, 30, 1102–1109.
Kuhlmann, M., & Mette, M. F. (2012). Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Molecular Biology, 79, 623–633.
Jiang, H., & Kohler, C. (2012). Evolution, function and regulation of genomic imprinting in plant seed development. Journal of Experimental Botany, 63, 4713–4722.
Berr, A., Shafiq, S., & Shen, W. H. (2011). Histone modifications in transcriptional activation during plant development. BBA Gene Regulatory Mechanisms, 1809, 567–576.
Wu, G., & Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development, 133, 3539–3547.
Lauter, N., Kampani, A., Carlson, S., Goebel, M., & Moose, S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proceedings of the National Academy of Sciences of the United States of America, 102, 9412–9417.
Kawashima, C. G., Yoshimoto, M., Maruyama-Nakashita, A., Tsuchiya, Y. N., Saito, K., Takahashi, H., & Dalmay, T. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its targets genes but in different cell types. The Plant Journal, 57, 313–321.
Allen, E., Xie, Z., Gustafson, A. M., & Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell, 121, 207–221.
Liang, G., Yang, F., & Yu, D. (2007). microRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. The Plant Journal, 62, 1046–1057.
Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., & Zhu, J. K. (2005). A miRNA involved in phosphate starvation response in Arabidopsis. Current Biology, 15, 2038–2043.
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., & Su, C. I. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell, 18, 412–421.
Zhou, Z. S., Zeng, H. Q., Liu, Z. P., & Yang, Z. M (2012). Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant, Cell and Environment, 35, 86–99.
Dubey, S., Saxena, S., Chauhan, A. S., Mathur, P., Rani, V., & Chakrabaroty, D. (2020). Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa). Environmental Science and Pollution Research, 27, 380–390.
Huang, S. Q., Xiang, A. L., Che, L. L., Chen, S., Li, H., Song, J. B., & Yang, Z. M. (2011). A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnology Journal, 8, 887–899.
Zhou, Z. S., Song, J. B., & Yang, Z. M. (2012). Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. Journal of Experimental Botany, 63, 4597–4613.
Lima, J. C., Arenhart, R. A., Margis-Pinheiro, M., & Margis, R. (2011). Aluminum triggers broad changes in microRNA expression in rice roots. Genetics and Molecular Research, 10, 2817–2832.
Ding, Y., Chen, Z., & Zhu, C. (2011). Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). Journal of Experimental Botany, 62, 3563–3573.
Zhou, M., Zheng, S., Liu, R., Lu, L., Zhang, C., Zhang, L., Yant, L., & Wu, Y. (2019). The genome-wide impact of cadmium on microRNA and mRNA expression in contrasting cd responsive wheat genotypes. BMC Genomics, 20, 615.
Carrasco-Gil, L., Álvarez-Fernández, A., Sobrino-Plata, J., Milán, R., Carpena-Ruiz, R. O., Leduc, D. L., Andrews, J. C., Abadía, J., & Hernández, L. E. (2011)complexation of hg with phytochelatins is important for plant hg tolerance. Plant, Cell & Environment, 34, 778–791.
Cobbett, C., & Goldsbrough, P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology, 53, 159–182.
Howden, R., Goldsbrough, P. B., Anderson, C. R., & Cobbett, C. S. (1995). Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiology, 107, 1059–1066.
Yamasaki, H., Abdel-Ghany, S. E., Bohu, C. M., Kobayashi, Y., Shikanai, T., & Pilon, M. (2007). Regulation of copper homeostasis but micro-RNA in Arabidopsis. Journal of Biological Chemistry, 282, 16369–16378.
Abdel-Ghany, S. E., & Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. Journal of Biological Chemistry, 283, 15932–11594.
Opdenakker, K., Remans, T., Keunen, E., Vangronsveld, J., & Cuypers, A. (2012). Exposure of Arabidopsis thaliana to Cd or Cu excess leads to oxidative stress mediated alterations in MAPKinase transcript levels. Environmental and Experimental Botany, 83, 53–61.
Chen, M., Meng, Y., Mao, C., Chen, D., & Wu, P. (2011). Methodological framework for functional characterization of plant microRNAs. Journal of Experimental Botany, 61, 2271–2280.
Sandmann, G., & Böger, P. (1980). Copper-mediated lipid peroxidation processes in photosynthetic membranes. Plant Physiology, 66, 797–800.
Maksymiec, W. (2004). Signaling responses in plants to heavy metal stress. Acta Physiologiae Plantarum, 29, 177–187.
Maksymiec, W., Wianowska, D., Dawidowicz, A. L., Radkiewicz, S., Mardarowicz, M., & Krupa, Z. (2005). The level of jasmonic acid in Arabidopsis thaliana and Phaseolus coccineus plants under heavy metal stress. Journal of Plant Physiology, 162, 1338–1346.
Peto, A., Lehotai, N., Lozano-Juste, J., León, J., Tari, I., Erdei, L., & Kolbert, Z. (2011). Involvement of nitric oxide and auxin signal transduction of copper-induced morphological responses in Arabidopsis seedlings. Annals of Botany, 108, 449–457.
Llave, C., Kasschau, K. D., Rector, M. A., & Carrington, J. C. (2002). Endogenous and silencing-associated small RNAs in plants. Plant Cell, 14, 1605–1619.
Zhang, H. Y., Xu, W. Z., Guo, J. B., He, Z. Y., & Ma, M. (2005). Coordinated responses of phytochelatins and metallothioneins to heavy metals in garlic seedlings. Plant Science, 169, 1059–1065.
Si-Ammour, A., Windels, D., Arn-Bouloires, E., Kutter, C., Ailhas, J., Meins, F., Jr, & Vazquez, F. (2011). MiR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiology, 157, 683–691.
Imtiaza, M., Mushtaqc, M. A., Nawazd, M. A., Ashrafe, M., Rizwanf, M. S., Mehmoodg, S., Rizwang, O. A. M., Virkh, M. S., Ijazj, Q. S. R., Androutsopoulosk, V. P., Tsatsakisk, A. D., & Colemanl, M. D. (2021). Physiological and anthocyanin Biosinthesys gene response induced by vanadium stress in mustard genotypes with distinct photosynthetic activity. Envirol Toxicol and Pharmacol. https://doi.org/10.1016/j.etap.2018.06.003
Dos Reis, S. P., Lima, A. M., & de Souza, C. R. B. (2012). Recent molecular advances on downstream plant responses to abiotic stress. International Journal of Molecular Sciences, 13, 8628–8647.
Verbruggen, N., Hermans, C., & Schat, H. (2009). Molecular mechanisms of metal hyper accumulation in plants. New Physiologist, 181(4), 759–776.
Kopriva, S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Annals of Botany, 97, 479–495.
Elobeid, M., Gobel, C., Feussner, I., & Polle, A. (2012). Cadmium interferes with auxin physiology and lignifications in poplar. Journal of Experimental Botany, 63, 1413–1421.
Marmiroli, M., Antonioli, G., Maestri, E., & Marmiroli, N. (2005). Evidence of the involvement of plant lingo-cellulosic structure in the sequestration of Pb: An X-ray spectroscopy-based analysis. Environmental Pollution, 134, 217–227.
Aina, R., Sgorbati, S., Santagostino, A., Labra, M., Ghiani, A., & Citterio, S. (2004). Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hamp. Physiologia Plantarum, 121, 472–480.
Wang, H., Zhang, X., Liu, J., Kiba, T., Woo, J., Ojo, T., Hafner, M., Tuschl, T., Chua, N. H., & Wang, X. J. (2006). Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. The Plant Journal, 67, 292–304.
Xie, Z., Kasschau, K. D., & Carrington, J. C. (2003). Negative feedback regulation of Dicer-like1 in Arabidopsis by miRNA-guided mRNA degradation. Current Biology, 13, 784–789.
Jones-Rhoades, M. W., & Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell, 14, 787–799.
Wu, L., Zhou, H., Zhang, Q., Zhang, J., Ni, F., Liu, C., & Qi, Y. (2010). DNA methylation mediated by a microRNA pathway. Molecular Cell, 38, 465–475.
Lee, Y., Kim, M., Han, J., Yeom, K. H., Lee, S., Baek, S. H., & Kim, V. N. (2004). MicroRNA genes are transcribed by polymerase II. The EMBO Journal, 23, 4051–4060.
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., Steward, R., & Chen, X. (2005). Methylation as a crucial step in plant miRNA biogenesis. Science, 307, 932–935.
Kurihara, Y., Takashi, Y., & Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212.
Gao, J., Luo, M., Peng, H., Chen, F. B., & Li, W. B. (2019). Characterization of cadmium responsive MicroRNAs and their target genes in maize (Zea mays) roots. BMC Molecular Biology, 20, 14.
Wang, B. X., Cheng, D., Chen, Z. Y., Zhang, M. M., Zhang, G. Q., Jiang, M. Y., & Tan, M. P. (2019). Bioinformatic exploration of the targets of Xylem Sap miRNAs in Maize under Cadmium stress. International Journal of Molecular Sciences, 20, 1474.
Chinnusamy, V., & Zhu, J. K. (2011). Epigenetic regulation of stress responses in plants. Current Opinion in Plant Biology, 212, 133–139.
Ranieri, E., Moustakas, K., Barbafieri, M., Ranieri, A. C., Herrera-Meli´ an, J. A., Petrella, A., et al. (2020). Phytoextraction technologies for mercury-and chromium-contaminated soil: A review. Journal of Chemical Technology and Biotechnology, 95, 317–327.
Ghosh, S., Adhikari, S., Adhikari, S., & Hossain, Z. (2022). Contribution of plant miRNA on studies towards understanding heavy metal stress responses: Current status and future perspectives. Environmental and Experimental Botany, 194, 104705.
Kumar, K., Shinde, A., Aeron, V., Verma, A., & Arif, N. S. (2022). Genetic engineering of plants for phytoremediation: Advances and challenges. Journal of Plant Biochemistry and Biotechnology, 12, 1–9.
Zhou, X., Wang, G., & Zhang, W. (2007). UV-B responsive microRNA genes in Arabidopsis thaliana. Molecular Systems Biology, 3(1), 103.
Nazir, R., Khan, M., Masab, M., Rehman, H. U., Rauf, N. U., Shahab, S., et al. (2015). Accumulation of heavy metals (Ni, Cu, Cd, Cr, Pb, Zn, Fe) in the soil, water and plants and analysis of physico-chemical parameters of soil and water collected from Tanda Dam Kohat. Journal of Pharmaceutical Sciences and Research, 7(3), 89.
Yang, Z., Yang, F., Liu, J. L., Wu, H. T., Yang, H., Shi, Y., Liu, J., Zhang, Y. F., Luo, Y. R., & Chen, K. M. (2022). Heavy metal transporters: Functional mechanisms, regulation, and application in phytoremediation. Science of the Total Environment, 809, 151099.
Kumar, K., Shinde, A., Aeron, V., et al. (2023). Genetic engineering of plants for phytoremediation: Advances and challenges. Journal of Plant Biochemistry and Biotechnology, 32, 12–30.
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., Wu, L., Li, S., Zhou, H., Long, C., et al. (2008). Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5’ terminal nucleotide. Cell, 133, 116–127.
Author information
Authors and Affiliations
Contributions
All the authors contributed equally.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
The authors give the consent for publication.
Conflict of Interest/Competing Interests
The authors do not have any conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Talukder, P., Saha, A., Roy, S. et al. Role of mi RNA in Phytoremediation of Heavy Metals and Metal Induced Stress Alleviation. Appl Biochem Biotechnol 195, 5712–5729 (2023). https://doi.org/10.1007/s12010-023-04599-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04599-3