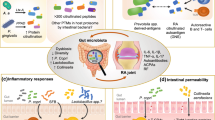
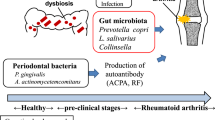
Abstract
Purpose of Review
Host-microbiome interactions have been implicated in the pathophysiology of rheumatoid arthritis (RA), but the data linking specific microbes to RA is largely associative. Here, we review recent studies that have interrogated specific mechanistic links between microbes and host in the setting of RA.
Recent Findings
Several candidate bacterial species and antigens that may trigger the conversion of an anti-bacterial to an autoimmune response have been recently identified. Additional studies have identified microbial metabolic pathways that are altered in RA. Some of these microbial species and metabolic pathways have been validated in mouse models to induce RA-like immune responses, providing initial evidence of specific mechanisms by which the microbiota contributes to the development of RA.
Summary
Several microbial species, antigens, and metabolites have been identified as potential contributors to RA pathophysiology. Further interrogation and validation of these pathways may identify novel biomarkers of or therapeutic avenues for RA.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10.
Malard F, Dore J, Gaugler B, Mohty M. Introduction to host microbiome symbiosis in health and disease. Mucosal Immunol. 2021;14(3):547–54.
Wilkins LJ, Monga M, Miller AW. Defining dysbiosis for a cluster of chronic diseases. Sci Rep. 2019;9(1):12918.
Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360: j5145.
Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202.
Dagar S, Singh J, Saini A, Kumar Y, Chhabra S, Minz RW, et al. Gut bacteriome, mycobiome and virome alterations in rheumatoid arthritis. Front Endocrinol. 2023;13:1044673.
Wang D-W, Pang X-T, Zhang H, Gao H-X, Leng Y-F, Chen F-Q, et al. Gut microbial dysbiosis in rheumatoid arthritis: A systematic review protocol of case-control studies. BMJ Open. 2022;12(4): e052021.
Attur M, Scher JU, Abramson SB, Attur M. Role of intestinal dysbiosis and nutrition in rheumatoid arthritis. Cells. 2022;11(15):2436.
Wilson TM, Trent B, Kuhn KA, Demoruelle MK. Microbial influences of mucosal immunity in rheumatoid arthritis. Curr Rheumatol Rep. 2020;22(11):83.
Konig MF. The microbiome in autoimmune rheumatic disease. Best Pract Res Clin Rheumatol. 2020;34(1): 101473.
Chriswell ME, Kuhn KA. Microbiota-mediated mucosal inflammation in arthritis. Best Pract Res Clin Rheumatol. 2019;33(6): 101492.
Holers VM, Demoruelle MK, Kuhn KA, Buckner JH, Robinson WH, Okamoto Y, et al. Rheumatoid arthritis and the mucosal origins hypothesis: Protection turns to destruction. Nat Rev Rheumatol. 2018;14(9):542–57.
Deane KD. Preclinical Rheumatoid Arthritis and Rheumatoid Arthritis Prevention. Curr Rheumatol Rep. 2018;20(8):50.
Romero V, Fert-Bober J, Nigrovic PA, Darrah E, Haque UJ, Lee DM, et al. Immune-mediated pore-forming pathways induce cellular hypercitrullination and generate citrullinated autoantigens in rheumatoid arthritis. Sci Transl Med. 2013;5(209):209ra150–1.
Gabarrini G, de Smit M, Westra J, Brouwer E, Vissink A, Zhou K, et al. The peptidylarginine deiminase gene is a conserved feature of Porphyromonas gingivalis. Sci Rep. 2015;5(1):13936.
Wegner N, Wait R, Sroka A, Eick S, Nguyen K-A, Lundberg K, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: Implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62(9):2662–72.
Konig MF, Abusleme L, Reinholdt J, Palmer RJ, Teles RP, Sampson K, et al. Aggregatibacter actinomycetemcomitans–induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med. 2016;8(369):369ra176-369ra1.
Möller B, Kollert F, Sculean A, Villiger PM. Infectious triggers in periodontitis and the gut in rheumatoid arthritis (RA): A complex story about association and causality. Front Immunol. 2020;11:1108.
Maeda Y, Takeda K. Host–microbiota interactions in rheumatoid arthritis. Exp Mol Med. 2019;51(12):1–6.
Gómez-Bañuelos E, Mukherjee A, Darrah E, Andrade F. Rheumatoid arthritis-associated mechanisms of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. J Clin Med. 2019;8(9):1309.
Clark RA. Fishing with autoantibodies nets a gut bacteria that drives arthritis. Sci Immunol. 2022;7(78):eadf9316.
•• Chriswell ME, Lefferts AR, Clay MR, Hsu AR, Seifert J, Feser ML, et al. Clonal IgA and IgG autoantibodies from individuals at risk for rheumatoid arthritis identify an arthritogenic strain of Subdoligranulum. Sci Transl Med. 2022;14(668):eabn5166 In this study, the authors utilize monoclonal antibodies derived from individuals at risk for and with RA to identify potentially cross-reactive bacteria and demonstrate that these antibody-bound bacteria are able to induce T cell responses in patients with RA as well as arthritis in mice.
•• Brewer RC, Lanz TV, Hale CR, Sepich-Poore GD, Martino C, Swafford AD, et al. Oral mucosal breaks trigger anti-citrullinated bacterial and human protein antibody responses in rheumatoid arthritis. Sci Transl Med. 2023;15(684):eabq8476. This study describes the presence of citrullinated oral bacteria in the circulation of patients with RA to which ACPA as well as associated anti-citrullinated bacteria antibodies could bind..
Moentadj R, Wang Y, Bowerman K, Rehaume L, Nel H, Cuiv PO, et al. Streptococcus species enriched in the oral cavity of patients with RA are a source of peptidoglycan-polysaccharide polymers that can induce arthritis in mice. Ann Rheum Dis. 2021;80(5):573–81.
Lim JJ, Jones CM, Loh TJ, Ting YT, Zareie P, Loh KL, et al. The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR. Sci Immunol. 2021;6(58):eabe0896.
Nguyen H, James EA. Immune recognition of citrullinated epitopes. Immunology. 2016;149(2):131–8.
Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis. 2019;78(5):590–3.
Abdelsalam NA, Hegazy SM, Aziz RK. The curious case of Prevotella copri. Gut Microbes. 2023;15(2):2249152.
Miyauchi E, Shimokawa C, Steimle A, Desai MS, Ohno H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat Rev Immunol. 2023;23(1):9–23.
Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis & Rheumatology. 2017;69(5):964–75.
Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, et al. Two rheumatoid arthritis–specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Investig. 2017;127(8):2946–56.
• Pianta A, Chiumento G, Ramsden K, Wang Q, Strle K, Arvikar S, et al. Identification of novel, immunogenic HLA–DR-presented Prevotella copri peptides in patients with rheumatoid arthritis. Arthritis Rheumatol. 2021;73(12):2200–5. This study identifies through mass spectrometry P. copri neoepitopes that stimulate T cells from patients with RA.
Wang H, Ong E, Kao JY, Sun D, He Y. Reverse microbiomics: A new reverse dysbiosis analysis strategy and its usage in prediction of autoantigens and virulent factors in dysbiotic gut microbiomes from rheumatoid arthritis patients. Front Microbiol. 2021;12:633732.
•• Nii T, Maeda Y, Motooka D, Naito M, Matsumoto Y, Ogawa T, et al. Genomic repertoires linked with pathogenic potency of arthritogenic Prevotella copri isolated from the gut of patients with rheumatoid arthritis. Ann Rheum Dis. 2023;82(5):621–9. In this study, the authors identify P. copri strains with a 100kbp transposon unique to the microbiome of individuals with RA, which could enhance arthritis in murine models.
Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol. 2021;39(1):449–79.
Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5(193):193ra91-ra91.
Pongratz G, Lowin T, Sewerin P, Zaucke F, Jenei-Lanzl Z, Pauly T, et al. Tryptophan metabolism in rheumatoid arthritis is associated with rheumatoid factor and predicts joint pathology evaluated by the rheumatoid arthritis MRI score (RAMRIS). Clin Exp Rheumatol. 2019;37(3):450–7.
Li J, Che N, Xu L, Zhang Q, Wang Q, Tan W, et al. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin Rheumatol. 2018;37(6):1493–502.
Labadarios D, McKenzie DY, Dickerson JWT, Parke DV. Metabolic abnormalities of tryptophan and nicotinic acid in patients with rheumatoid arthritis. Rheumatology. 1978;17(4):227–32.
Schroecksnadel K, Kaser S, Ledochowski M, Neurauter G, Mur E, Herold M, et al. Increased degradation of tryptophan in blood of patients with rheumatoid arthritis. J Rheumatol. 2003;30(9):1935–9.
Forrest CM, Kennedy A, Stone TW, Stoy N, Darlington LG. Kynurenine and neopterin levels in patients with rheumatoid arthritis and osteoporosis during drug treatment. Adv Exp Med Biol. 2003;527:287–95.
Yu D, Du J, Pu X, Zheng L, Chen S, Wang N, et al. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front Cell Infect Microbiol. 2022;11:763507.
• Luo Y, Tong Y, Wu L, Niu H, Li Y, Su LC, et al. Alteration of gut microbiota in individuals at high-risk for rheumatoid arthritis associated with disturbed metabolome and the initiation of arthritis through the triggering of mucosal immunity imbalance. Arthritis Rheumatol. 2023;75(10):1736–48. This study describes multiple changes in the circulating metabolome of patients with RA, including that of the tryptophan pathway.
•• Seymour BJ, Trent B, Allen B, Berlinberg AJ, Tangchittsumran J, Jubair WK, et al. Microbiota-dependent indole production stimulates the development of collagen-induced arthritis in mice. J Clin Invest. 2023. https://doi.org/10.1172/JCI167671. Microbial dysbiosis in this study was associated with tryptophan catabolism to indole, which resulted in increased arthritis severity, Th17 cell expansion, and pathogenic autoantibody formation in collagen-induced arthritis.
Berlinberg AJ, Regner EH, Stahly A, Brar A, Reisz JA, Gerich ME, et al. Multi ‘omics analysis of intestinal tissue in ankylosing spondylitis identifies alterations in the tryptophan metabolism pathway. Front Immunol. 2021;12:587119.
Choi S-C, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med. 2020;12(551):eaax2220.
Sonner JK, Keil M, Falk-Paulsen M, Mishra N, Rehman A, Kramer M, et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat Commun. 2019;10(1):4877.
Montgomery TL, Eckstrom K, Lile KH, Caldwell S, Heney ER, Lahue KG, et al. Lactobacillus reuteri tryptophan metabolism promotes host susceptibility to CNS autoimmunity. Microbiome. 2022;10(1):198.
Brown J, Robusto B, Morel L. Intestinal dysbiosis and tryptophan metabolism in autoimmunity. Front Immunol. 2020;11:1741.
Moulin D, Millard M, Taïeb M, Michaudel C, Aucouturier A, Lefèvre A, et al. Counteracting tryptophan metabolism alterations as a new therapeutic strategy for rheumatoid arthritis. Ann Rheum Dis. 2023. https://doi.org/10.1136/ard-2023-224014.
He J, Chu Y, Li J, Meng Q, Liu Y, Jin J, et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci Adv. 2022;8(6):eabm1511.
Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. 2020;31(4):837-51.e10.
Jiang L, Shang M, Yu S, Liu Y, Zhang H, Zhou Y, et al. A high-fiber diet synergizes with Prevotella copri and exacerbates rheumatoid arthritis. Cell Mol Immunol. 2022;19(12):1414–24.
•• Hong M, Li Z, Liu H, Zheng S, Zhang F, Zhu J, et al. Fusobacterium nucleatum aggravates rheumatoid arthritis through FadA-containing outer membrane vesicles. Cell Host Microbe. 2023;31(5):798–810.e7. In this study, outer membrane vesicles from F. nucleatum were identified in the synovial fluid of individuals with RA, and colonization of mice with F. nucleatum could enhance collagen-induced arthritis.
Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 2018;8(1):14305.
• Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. 2020;11(1):1995. This study describes increased intestinal permeability in individuals at risk for and with RA, and restoring intestinal homeostasis reduces arthritis severity in collagen-induced arthritis.
Brandl C, Bucci L, Schett G, Zaiss MM. Crossing the barriers: Revisiting the gut feeling in rheumatoid arthritis. Eur J Immunol. 2021;51(4):798–810.
Funding
BJS received funding through the National Institute of Allergy and Infectious Diseases grants F30AI174817 and T32AI007405. KAK received funding through the National Institute of Allergy and Infectious Diseases grant U01AI101981 and the National Institute of Arthritis and Musculoskeletal and Skin Diseases grants R01AR075933 and P30AR079369.
Author information
Authors and Affiliations
Contributions
BJS, BEA, and KAK wrote the manuscript text, and BJS prepared Fig. 1. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Seymour, B.J., Allen, B.E. & Kuhn, K.A. Microbial Mechanisms of Rheumatoid Arthritis Pathogenesis. Curr Rheumatol Rep 26, 124–132 (2024). https://doi.org/10.1007/s11926-024-01135-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11926-024-01135-y