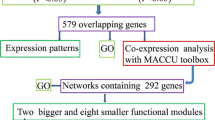
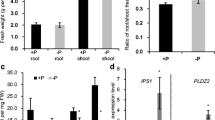
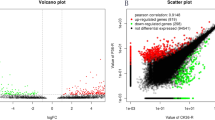
Abstract
Phosphate (Pi) is one of the major nutrients for crop growth and yield. Although several studies have revealed a global view of Pi starvation responses in rice, the detailed features were not well-addressed. To identify differentially expressed genes associated with phosphate starvation on a genome scale, we analyzed transcriptome data of roots from 2-week old seedlings that were grown on Pi-sufficient or -deficient media for 7 and 21 days. Using publicly available RNA-sequencing data, we subsequently identified 820 up-regulated genes in roots under Pi starvation. Gene ontology enrichment analysis of these genes indicated that secondary metabolic processes are most significantly enriched under Pi starvation, and Pi transport and defense to biotic stress also play significant roles in response against Pi deficiency. Functional classification analysis using MapMan emphasizes the significance of transcription factors, such as MYB, WRKY, and bHLH, various transporters, and genes in secondary metabolic processes. Use of promoter trap lines or transgenic plants expressing the GUS reporter gene under the control of Pi starvation-inducible gene promoters confirmed the meta-expression patterns of two genes stimulated by Pi starvation, suggesting novel promoters for enhancing Pi use efficiency. In addition to the identification of two novel promoters for Pi starvation response, cis-regulatory elements for the regulation of Pi starvation are suggested. Overall, our study provides a global view of Pi starvation response based on transcriptome data and novel tools for improving PUE and Pi uptake in rice, a model crop plant.







Similar content being viewed by others
References
Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, Xu G (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57:798–809
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51:1019–1029
Arite T, Kameoka H, Kyozuka J (2012) Strigolactone positively controls crown root elongation in rice. J Plant Growth Regul 31:165
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:e39–e49
Bao Z, Watanabe A, Sasaki K, Okubo T, Tokida T, Liu D, Ikeda S, Imaizumi-Anraku H, Asakawa S, Sato T, Mitsui H, Minamisawa K (2014) A rice gene for microbial symbiosis, Oryza sativa CCaMK, reduces CH4 Flux in a paddy field with low nitrogen input. Appl Environ Microbiol 80:995–2003
Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6:e1001102
Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59:2479–2497
Cao P, Jung K-H, Choi D, Hwang D, Zhu J, Ronald PC (2012) The rice oligonucleotide array database: an atlas of rice gene expression. Rice 5:17
Chandran AKN, Lee GS, Yoo YH, Yoon UH, Ahn BO, Yun DW, Kim JH, Choi HK, An G, Kim TH, Jung KH (2016) Functional classification of rice flanking sequence tagged genes using MapMan terms and global understanding on metabolic and regulatory pathways affected by dxr mutant having defects in light response. Rice 9:17
Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC (2016) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res 44:D1154–D1160
Cordell D, Rosemarin A, Schroder JJ, Smit AL (2011) Towards global phosphorus security: a systems framework for phosphorus recovery and reuse options. Chemosphere 84:747–758
Dai X, Wang Y, Zhang WH (2016) OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J Exp Bot 67:947–960
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, Bouwmeester H, Becard G, Beveridge CA, Rameau C, Rochange SF (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194
Hatorangan MR, Sentausa E, Wijaya GY (2009) In silico identification of cis-regulatory elements of phosphate transporter genes in rice (Oryza sativa L.). J Crop Sci Biotechnol 12:25–30
Higo K, Ugawa Y, Iwamoto M, Higo H (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26:358–359
Hirsch J, Marin E, Floriani M, Chiarenza S, Richaud P, Nussaume L, Thibaud MC (2006) Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88:1767–1771
Jefferson RA (1989) The GUS reporter gene system. Nature 342:837–838
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156:1164–1175
Jung KH, Dardick C, Bartley LE, Cao P, Phetsom J, Canlas P, Seo YS, Shultz M, Ouyang S, Yuan Q, Frank BC, Ly E, Zheng L, Jia Y, Hsia AP, An K, Chou HH, Rocke D, Lee GC, Schnable PS, An G, Buell CR, Ronald PC (2008) Refinement of light-responsive transcript lists using rice oligonucleotide arrays: evaluation of gene-redundancy. PLoS One. https://doi.org/10.1371/journal.pone.0003337
Juszczuk IM, Wiktorowska A, Malusá E, Rychter AM (2004) Changes in the concentration of phenolic compounds and exudation induced by phosphate deficiency in bean plants (Phaseolus vulgaris L.). Plant Soil 267:41–49
Kapulnik Y, Delaux PM, Resnick N, Mayzlish-Gati E, Wininger S, Bhattacharya C, Sejalon-Delmas N, Combier JP, Becard G, Belausov E, Beeckman T, Dor E, Hershenhorn J, Koltai H (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233:209–216
Kato-Noguchi H (2011) Convergent or parallel molecular evolution of momilactone A and B: potent allelochemicals, momilactones have been found only in rice and the moss Hypnum plumaeforme. J Plant Physiol 168:1511–1516
Kim SY, Madrid AV, Park ST, Yang SJ, Olofsdotter M (2005) Evaluation of rice allelopathy inhydroponics. Weed Res 45:74–79
Kim SR, Lee DY, Yang JI, Moon SO, An G (2009) Cloning vectors for rice. J Plant Biol 52:73
Koevoets IT, Venema JH, Elzenga JT, Testerink C (2016) Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front Plant Sci 7:1335
Koltai H, Kapulnik Y (2011) Strigolactones as mediators of plant growth responses to environmental conditions. Plant Signal Behav 6:37–41
Kong CH, Li HB, Hu F, Xu XH, Wang P (2006) Allelochemicals released by rice roots and residues in soil. Plant Soil 288:47–56
Koyama T, Ono T, Shimizu M, Jinbo T, Mizuno R, Tomita K, Mitsukawa N, Kawazu T, Kimura T, Ohmiya K, Sakka K (2005) Promoter of Arabidopsis thaliana phosphate transporter gene drives root-specific expression of transgene in rice. J Biosci Bioeng 99:38–42
Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483:341–344
Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H (2012) Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol 160:319–331
Kumar M, Pandya-Kumar N, Kapulnik Y, Koltai H (2015) Strigolactone signaling in root development and phosphate starvation. Plant Signal Behav 10:e1045174
Lapis-Gaza HR, Jost R, Finnegan PM (2014) Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14:334
Lee SH, Shon YG, Lee SI, Kim CY, Koo JC, Lim CO, Choi YJ, Han CD, Chung CH, Choe ZR, Cho MJ (1999) Cultivar variability in the Agrobacterium-rice cell interaction and plant regeneration. Physiol Plant 107:338–340
Lee DK, Chung PJ, Jeong JS, Jang G, Bang SW, Jung H, Kim YS, Ha SH, Choi YD, Kim JK (2017) The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol J 15:754–764
Li L, Qiu X, Li X, Wang S, Lian X (2009) The expression profile of genes in rice roots under low phosphorus stress. Sci China C Life Sci 52:1055–1064
Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, Yang WC, Hooiveld GJ, Charnikhova T, Bouwmeester HJ, Bisseling T, Geurts R (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23:3853–3865
Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, Doumas P, Nacry P, Herrerra-Estrella L, Nussaume L, Thibaud MC (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. PNAS 102:11934–11939
Morris RT, O’Connor TR, Wyrick JJ (2008) Osiris: an integrated promoter database for Oryza sativa L. Bioinformatics 24:2915–2917
Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. PNAS 93:10519–10523
Mudge SR, Smith FW, Richardson AE (2003) Root-specific and phosphate-regulated expression of phytase under the control of a phosphate transporter promoter enables Arabidopsis to grow on phytate as a sole P source. Plant Sci 165:871–878
Nagy R, Vasconcelos MJ, Zhao S, McElver J, Bruce W, Amrhein N, Raghothama KG, Bucher M (2006) Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant Biol (Stuttg) 8:186–197
Nilsson L, Muller R, Nielsen TH (2010) Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol Plant 139:129–143
Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK (2006) Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J Exp Bot 57:2867–2878
Oono Y, Kawahara Y, Kanamori H, Mizuno H, Yamagata H, Yamamoto M, Hosokawa S, Ikawa H, Akahane I, Zhu Z, Wu J, Itoh T, Matsumoto T (2011) mRNA-Seq reveals a comprehensive transcriptome profile of rice under phosphate stress. Rice 4:50–65
Oono Y, Kawahara Y, Yazawa T, Kanamori H, Kuramata M, Yamagata H, Hosokawa S, Minami H, Ishikawa S, Wu J, Antonio B, Handa H, Itoh T, Matsumoto T (2013) Diversity in the complexity of phosphate starvation transcriptomes among rice cultivars based on RNA-Seq profiles. Plant Mol Biol 83:523–537
Oropeza-Aburto A, Cruz-Ramirez A, Acevedo-Hernandez GJ, Perez-Torres CA, Caballero-Perez J, Herrera-Estrella L (2012) Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J Exp Bot 63:2189–2202
Raghothama KG, Karthikeyan AS (2005) Phosphate acquisition. Plant Soil 274:37
Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116(2):447–453
Schroder JJ, Smit AL, Cordell D, Rosemarin A (2011) Improved phosphorus use efficiency in agriculture: a key requirement for its sustainable use. Chemosphere 84:822–831
Schunmann PH, Richardson AE, Vickers CE, Delhaize E (2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136:4205–4214
Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25:4285–4304
Shen L, Lin W (2007) Effects of phosphorus levels on allelopathic potential of rice cocultured with barnyardgrass. Allelopathy J 19:393–402
Shiono K, Ando M, Nishiuchi S, Takahashi H, Watanabe K, Nakamura M, Matsuo Y, Yasuno N, Yamanouchi U, Fujimoto M, Takanashi H, Ranathunge K, Franke RB, Shitan N, Nishizawa NK, Takamure I, Yano M, Tsutsumi N, Schreiber L, Yazaki K, Nakazono M, Kato K (2014) RCN1/OsABCG5, an ATP-binding cassette (ABC) transporter, is required for hypodermal suberization of roots in rice (Oryza sativa). Plant J 80:40–51
Song B, Xiong J, Fang C, Qiu L, Lin R, Liang Y (2008) Allelopathic enhancement and differential gene expression in rice under low nitrogen treatment. J Chem Ecol 34:688–695
Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G (2014) Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot 65:6735–6746
Tseng IC, Hong CY, Yu SM, Ho TH (2013) Abscisic acid- and stress-induced highly proline-rich glycoproteins regulate root growth in rice. Plant Physiol 163:118–134
Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51:1118–1126
Urbanczyk-Wochniak E, Usadel B, Thimm O, Nunes-Nesi A, Carrari F, Davy M, Blasing O, Kowalczyk M, Weicht D, Polinceusz A, Meyer S, Stitt M, Fernie AR (2006) Conversion of MapMan to allow the analysis of transcript data from Solanaceous species: effects of genetic and environmental alterations in energy metabolism in the leaf. Plant Mol Biol 60:773–792
Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, Scheible WR, Gibon Y, Morcuende R, Weicht D, Meyer S, Stitt M (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138:1195–1204
Wang L, Liu D (2012) Arabidopsis purple acid phosphatase 10 is a component of plant adaptive mechanism to phosphate limitation. Plant Signal Behav 7:306–310
Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P (2009) Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol 51:663–674
Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, Shou H, Mo X, Mao C, Wu P (2014) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. PNAS 111:14953–14958
Xi L, Wen C, Fang S, Chen X, Nie J, Chu J, Yuan C, Yan C, Ma N, Zhao L (2015) Impacts of strigolactone on shoot branching under phosphate starvation in chrysanthemum (Dendranthema grandiflorum cv. Jinba). Front Plant Sci 6:694
Yamamoto E, Yonemaru J, Yamamoto T, Yano M (2012) OGRO: the overview of functionally characterized Genes in Rice online database. Rice (N Y) 5:26
Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P (2005) OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol 138:2087–2096
Yokota K, Soyano T, Kouchi H, Hayashi M (2010) Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice. Plant Cell Physiol 51:1436–1442
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice, vol 3. IRRI, Los Banos, pp 27–34
Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. PNAS 99:5732–5737
Zhang L, Hu B, Li W, Che R, Deng K, Li H, Yu F, Ling H, Li Y, Chu C (2014) OsPT2, a phosphate transporter, is involved in the active uptake of selenite in rice. New Phytol 201:1183–1191
Zhao H, Li H, Kong C, Xu X, Liang W (2005) Chemical response of allelopathic rice seedlings under varying environmental conditions. Allelopathy J 15:105–110
Acknowledgements
This work was supported by grants from the Next-Generation BioGreen 21 Program (PJ01100401 to KHJ), and Research Program for Agriculture Science & Technology Development (Project title: Construction of forage rice genetic population for adaptation to reclaimed land, Project No. PJ01358001 to HMP), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gho, YS., An, G., Park, HM. et al. A systemic view of phosphate starvation-responsive genes in rice roots to enhance phosphate use efficiency in rice. Plant Biotechnol Rep 12, 249–264 (2018). https://doi.org/10.1007/s11816-018-0490-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-018-0490-y