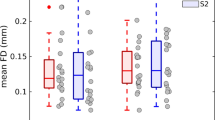
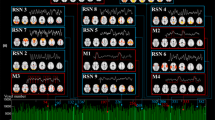
Abstract
Large-scale consortium efforts such as Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) and other collaborative efforts show that combining statistical data from multiple independent studies can boost statistical power and achieve more accurate estimates of effect sizes, contributing to more reliable and reproducible research. A meta- analysis would pool effects from studies conducted in a similar manner, yet to date, no such harmonized protocol exists for resting state fMRI (rsfMRI) data. Here, we propose an initial pipeline for multi-site rsfMRI analysis to allow research groups around the world to analyze scans in a harmonized way, and to perform coordinated statistical tests. The challenge lies in the fact that resting state fMRI measurements collected by researchers over the last decade vary widely, with variable quality and differing spatial or temporal signal-to-noise ratio (tSNR). An effective harmonization must provide optimal measures for all quality data. Here we used rsfMRI data from twenty-two independent studies with approximately fifty corresponding T1-weighted and rsfMRI datasets each, to (A) review and aggregate the state of existing rsfMRI data, (B) demonstrate utility of principal component analysis (PCA)-based denoising and (C) develop a deformable ENIGMA EPI template based on the representative anatomy that incorporates spatial distortion patterns from various protocols and populations.








Similar content being viewed by others
References
Adams, H. H., Hibar, D. P., Chouraki, V., Stein, J. L., Nyquist, P. A., Renteria, M. E., et al. (2016). Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nature Neuroscience, 19(12), 1569–1582. https://doi.org/10.1038/nn.4398.
Avants, B. B., Epstein, C. L., Grossman, M., & Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis, 12(1), 26–41.
Avants, B. B., Yushkevich, P., Pluta, J., Minkoff, D., Korczykowski, M., Detre, J., & Gee, J. C. (2010). The optimal template effect in hippocampus studies of diseased populations. Neuroimage, 49(3), 2457–2466.
Avants, B. B., Johnson, H. J., & Tustison, N. J. (2015). Neuroinformatics and the the insight ToolKit. Frontiers in Neuroinformatics, 9, 5. https://doi.org/10.3389/fninf.2015.00005.
Bajaj, S., Butler, A. J., Drake, D., & Dhamala, M. (2015a). Brain effective connectivity during motor-imagery and execution following stroke and rehabilitation. Neuroimage Clinical, 8, 572–582. https://doi.org/10.1016/j.nicl.2015.06.006.
Bajaj, S., Butler, A. J., Drake, D., & Dhamala, M. (2015b). Functional organization and restoration of the brain motor-execution network after stroke and rehabilitation. Frontiers in Human Neuroscience, 9, 173. https://doi.org/10.3389/fnhum.2015.00173.
Bajaj, S., Adhikari, B. M., Friston, K. J., & Dhamala, M. (2016). Bridging the gap: dynamic causal modeling and granger causality analysis of resting state functional magnetic resonance imaging. Brain Connectivity. https://doi.org/10.1089/brain.2016.0422.
Baloch, S., & Davatzikos, C. (2009). Morphological appearance manifolds in computational anatomy: groupwise registration and morphological analysis. Neuroimage, 45(S1), S73–S85.
Bearden, C. E., van Erp, T. G. M., Dutton, R. A., Tran, H., Zimmermann, L., Sun, D., et al. (2007). Mapping cortical thickness in children with 22q11.2 deletions. Cerebral Cortex, 17, 1889–1898.
Beckmann, C. F., & Smith, S. M. (2004). Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Transactions on Medical Imaging, 23(2), 137–152.
Beg, M. F., & Khan, A. (2007). Symmetric data attachment terms for large deformation image registration. IEEE Transactions on Medical Imaging, 26, 1179–1189.
Bis, J. C., DeCarli, C., Smith, A. V., van der Lijn, F., Crivello, F., Fornage, M., Debette, S., Shulman, J. M., Schmidt, H., Srikanth, V., Schuur, M., Yu, L., Choi, S. H., Sigurdsson, S., Verhaaren, B. F., DeStefano, A., Lambert, J. C., Jack CR Jr, Struchalin, M., Stankovich, J., Ibrahim-Verbaas, C. A., Fleischman, D., Zijdenbos, A., den Heijer, T., Mazoyer, B., Coker, L. H., Enzinger, C., Danoy, P., Amin, N., Arfanakis, K., van Buchem, M., de Bruijn, R. F., Beiser, A., Dufouil, C., Huang, J., Cavalieri, M., Thomson, R., Niessen, W. J., Chibnik, L. B., Gislason, G. K., Hofman, A., Pikula, A., Amouyel, P., Freeman, K. B., Phan, T. G., Oostra, B. A., Stein, J. L., Medland, S. E., Vasquez, A. A., Hibar, D. P., Wright, M. J., Franke, B., Martin, N. G., Thompson, P. M., Enhancing Neuro Imaging Genetics through Meta-Analysis Consortium, Nalls, M. A., Uitterlinden, A. G., Au, R., Elbaz, A., Beare, R. J., van Swieten, J., Lopez, O. L., Harris, T. B., Chouraki, V., Breteler, M. M., de Jager, P. L., Becker, J. T., Vernooij, M. W., Knopman, D., Fazekas, F., Wolf, P. A., van der Lugt, A., Gudnason, V., Longstreth WT Jr, Brown, M. A., Bennett, D. A., van Duijn, C., Mosley, T. H., Schmidt, R., Tzourio, C., Launer, L. J., Ikram, M. A., Seshadri, S., & Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. (2012). Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nature Genetics, 44(5), 545–551. https://doi.org/10.1038/ng.2237.
Biswal, B., Yetkin, F. Z., Haughton, V. M., & Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine, 34, 537–541.
Bodurka, J., Ye, F., Petridou, N., Murphy, K., & Bandettini, P. A. (2007). Mapping the MRI voxel volume in which thermal noise matches physiological noise-implications for fMRI. Neuroimage, 34(2), 542–549.
Boedhoe, P. S., Schmaal, L., Abe, Y., Ameis, S. H., Arnold, P. D., Batistuzzo, M. C., et al. (2016). Distinct subcortical volume alterations in pediatric and adult OCD: a worldwide meta- and mega-analysis. American Journal of Psychiatry. https://doi.org/10.1176/appi.ajp.2016.16020201.
Brooks, J. C., Faull, O. K., Pattinson, K. T., & Jenkinson, M. (2013). Physiological noise in brainstem FMRI. Frontiers in Human Neuroscience, 7, 623. https://doi.org/10.3389/fnhum.2013.00623.
Bustin, S. A. (2014). The reproducibility of biomedical research: sleepers awake! Biomolecular Detection and Quantification, 2, 35–42. https://doi.org/10.1016/j.bdq.2015.01.002.
Button, K. S., Ioannidis, J. P., Mokrysz, C., Nosek, B. A., Flint, J., Robinson, E. S., et al. (2013). Power failure: why small sample size undermines the reliability of neuroscience. Nature Reviews. Neuroscience, 14(5), 365–376. https://doi.org/10.1038/nrn3475.
Calhoun, V. D., & Adali, T. (2012). Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Reviews in Biomedical Engineering, 5, 60–73.
Calhoun, V. D., Wager, T. D., Krishnan, A., Rosch, K. S., Seymour, K. E., Nebel, M. B., Mostofsky, S. H., Nyalakanai, P., & Kiehl, K. (2017). Normalization of fMRI data using T1 versus EPI. Human Brain Mapping, 38(11), 5331–5342.
Çetin, M. S., Christensen, F., Abbott, C. C., Stephen, J. M., Mayer, A. R., Cañive, J. M., et al. (2014). Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage, 15(97), 117–126.
Chen, M., Lu, W., Chen, Q., Ruchala, K. J., & Olivera, G. H. (2008). A simple fixed-point approach to invert a deformation field. Medical Physics, 35, 81–88.
Cheung, M. R., & Krishnan, K. (2009). Interactive deformation registration of endorectal prostate mri using itk thin plate splines. Academic Radiology, 16, 351–357.
Choudhury, S., Fishman, J. R., McGowan, M. L., & Juengst, E. T. (2014). Big data, open science and the brain: lessons learned from genomics. Frontiers in Human Neuroscience, 8, 239. https://doi.org/10.3389/fnhum.2014.00239.
Collins, D., Holmes, C., Peters, T., & Evans, A. (1995). Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping, 3(3), 190–208.
Cusack, R., Brett, M., & Osswald, K. (2003). An evaluation of the use of magnetic field maps to undistort echo-planar images. Neuroimage, 18, 127–142.
Du, Y., Allen, E. A., He, H., Sui, J., Wu, L., & Calhoun, V. D. (2016). Artifact removal in the context of group ICA: a comparison of single-subject and group approaches. Human Brain Mapping, 37(3), 1005–1025. https://doi.org/10.1002/hbm.23086.
Fedorov, A., Li, X., Pohl, K. M., Bouix, S., Styner, M., Addicott, M., Wyatt, C., Daunais, J. B., Wells, W. M., & Kikinis, R. (2011). Atlas-guided segmentation of vervet monkey brain MRI. Open Neuroimaging Journal, 5, 186–197.
Fox, M. D., & Raichle, M. E. (2007). Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nature Reviews. Neuroscience, 8(9), 700–711. https://doi.org/10.1038/nrn2201.
Geng, X., Christensen, G. E., Gu, H., Ross, T. J., & Yang, Y. (2009). Implicit reference-based group-wise image registration and its application to structural and functional MRI. Neuroimage, 47(4), 1341–1351.
Gonzalez-Castillo, J., Handwerker, D. A., Robinson, M. E., Hoy, C. W., Buchanan, L. C., Saad, Z. S., & Bandettini, P. A. (2014). The spatial structure of resting state connectivity stability on the scale of minutes. Frontiers in Neuroscience, 8, 138. https://doi.org/10.3389/fnins.2014.00138.
Greve, D. N., Mueller, B. A., Liu, T., Turner, J. A., Voyvodic, J., Yetter, E., Diaz, M., McCarthy, G., Wallace, S., Roach, B. J., Ford, J. M., Mathalon, D. H., Calhoun, V. D., Wible, C. G., Brown, G. G., Potkin, S. G., & Glover, G. (2011). A novel method for quantifying scanner instability in fMRI. Magnetic Resonance in Medicine, 65(4), 1053–1061. https://doi.org/10.1002/mrm.22691.
Guadalupe, T., Mathias, S. R., vanErp, T. G., Whelan, C. D., Zwiers, M. P., Abe, Y., et al. (2017). Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging and Behavior, 11(5), 1497–1514. https://doi.org/10.1007/s11682-016-9629-z.
Gusnard, D. A., & Raichle, M. E. (2001). Searching for a baseline: functional imaging and the resting human brain. Nature Reviews. Neuroscience, 2(10), 685–694.
Hayasaka, S. (2013). Functional connectivity networks with and without global signal correction. Frontiers in Human Neuroscience, 7, 880. https://doi.org/10.3389/fnhum.2013.00880.
Heiland, S. (2008). From A as in aliasing to Z as in zipper: artifacts in MRI. Clinical Neuroradiology, 18, 25–36.
Hibar, D. P., Stein, J. L., Renteria, M. E., Arias-Vasquez, A., Desrivieres, S., Jahanshad, N., et al. (2015). Common genetic variants influence human subcortical brain structures. Nature, 520(7546), 224–229. https://doi.org/10.1038/nature14101.
Hibar, D. P., Westlye, L. T., van Erp, T. G., Rasmussen, J., Leonardo, C. D., Faskowitz, J., et al. (2016). Subcortical volumetric abnormalities in bipolar disorder. Molecular Psychiatry, 21(12), 1710–1716. https://doi.org/10.1038/mp.2015.227.
Hibar, D. P., Adams, H. H., Jahanshad, N., Chauhan, G., Stein, J. L., Hofer, E., et al. (2017). Novel genetic loci associated with hippocampal volume. Nature Communications, 8, 13624. https://doi.org/10.1038/ncomms13624.
Hoogman, M., Bralten, J., Hibar, D. P., Mennes, M., Zwiers, M. P., Schweren, L. S., et al. (2017). Subcortical brain volume differences in participants with attention deficit hyperactivity disorder in children and adults: a cross-sectional mega-analysis. Lancet Psychiatry, 4(4), 310–319.
Hutton, C., Bork, A., Josephs, O., Deichmann, R., Ashburner, J., & Turner, R. (2002). Image distortion correction in fMRI: a quantitative evaluation. Neuroimage, 16, 217–240.
Ibanez, L., Ng, L., Gee, J., & Aylward, S. (2002). Registration patterns: the generic framework for image registration of the insight toolkit. Proceedings of IEEE International Symposium on Biomedical Imaging, 345–348.
Ikram, M. A., Fornage, M., Smith, A. V., Seshadri, S., Schmidt, R., Debette, S., Vrooman, H. A., Sigurdsson, S., Ropele, S., Taal, H. R., Mook-Kanamori, D. O., Coker, L. H., Longstreth WT Jr, Niessen, W. J., DeStefano, A., Beiser, A., Zijdenbos, A. P., Struchalin, M., Jack CR Jr, Rivadeneira, F., Uitterlinden, A. G., Knopman, D. S., Hartikainen, A. L., Pennell, C. E., Thiering, E., Steegers, E. A., Hakonarson, H., Heinrich, J., Palmer, L. J., Jarvelin, M. R., McCarthy, M., Grant, S. F., St Pourcain, B., Timpson, N. J., Smith, G. D., Sovio, U., Early Growth Genetics Consortium, Nalls, M. A., Au, R., Hofman, A., Gudnason, H., van der Lugt, A., Harris, T. B., Meeks, W. M., Vernooij, M. W., van Buchem, M., Catellier, D., Jaddoe, V. W., Gudnason, V., Windham, B. G., Wolf, P. A., van Duijn, C., Mosley TH Jr, Schmidt, H., Launer, L. J., Breteler, M. M., DeCarli, C., & Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. (2012). Common variants at 6q22 and 17q21 are associated with intracranial volume. Nature Genetics, 44(5), 539–544. https://doi.org/10.1038/ng.2245.
Jahanshad, N., Kochunov, P. V., Sprooten, E., Mandl, R. C., Nichols, T. E., Almasy, L., Blangero, J., Brouwer, R. M., Curran, J. E., de Zubicaray, G. I., Duggirala, R., Fox, P. T., Hong, L. E., Landman, B. A., Martin, N. G., McMahon, K. L., Medland, S. E., Mitchell, B. D., Olvera, R. L., Peterson, C. P., Starr, J. M., Sussmann, J. E., Toga, A. W., Wardlaw, J. M., Wright, M. J., Hulshoff Pol, H. E., Bastin, M. E., McIntosh, A. M., Deary, I. J., Thompson, P. M., & Glahn, D. C. (2013). Multi-site genetic analysis of diffusion images and voxelwise heritability analysis: a pilot project of the ENIGMA-DTI working group. Neuroimage, 81, 455–469.
Jahanshad, N., Roshchupkin, G., Faskowitz, J., Hibar, D. P., Gutman, B. A., Adams, A. H. H., et al. (2018). Multisite metaanalysis of image-wide genome-wide associations with morphometry (Imaging genetics ). Elsvier Inc.
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W., & Smith, S. M. (2012). FSL. Neuroimage, 62, 782–790.
Jezzard, P. (2012). Correction of geometric distortion in fMRI data. Neuroimage, 62(2), 648–651. https://doi.org/10.1016/j.neuroimage.2011.09.010.
Kikinis, R., & Pieper, S. (2011). 3d slicer as a tool for interactive brain tumor segmentation. Conference Proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society, 6982–6984.
Klein, A., Andersson, J., Ardekani, B. A., Ashburner, J., Avants, B., Chiang, M. C., Christensen, G. E., Collins, D. L., Gee, J., Hellier, P., Song, J. H., Jenkinson, M., Lepage, C., Rueckert, D., Thompson, P., Vercauteren, T., Woods, R. P., Mann, J. J., & Parsey, R. V. (2009). Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage, 46(3), 786–802. https://doi.org/10.1016/j.neuroimage.2008.12.037.
Kochunov, P., Lancaster, J. L., Thompson, P., Woods, R., Mazziotta, J., Hardies, J., & Fox, P. (2001). Regional spatial normalization: Toward an optimal target. Journal of Computer Assisted Tomography, 25(5), 805–816.
Kruger, G., & Glover, G. H. (2001). Physiological noise in oxygenation- sensitive magnetic resonance imaging. Magnetic Resonance in Medicine, 46, 631–637.
Lancaster, J. L., Fox, P., Downs, H., Nickerson, D., Hander, T., Mallah, M., et al. (1999). Global spatial normalization of the human brain using convex hulls. Journal of Nuclear Medicine, 40(6), 942–955.
Major Depressive Disorder Working Group of the Psychiatric, G. C, Ripke, S., Wray, N. R., Lewis, C. M., Hamilton, S. P., Weissman, M. M., et al. (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry, 18(4), 497–511. https://doi.org/10.1038/mp.2012.21.
Marchenko, V. A., & Pastur, L. A. (1967). Distribution of eigenvalues for some sets of random matrices. Mathematics of the USSR-Sbornik, 1(4).
Margulies, D. S., Clare Kellya, A. M., Uddin, L. Q., Biswal, B. B., Xavier Castellanos, F., & Milham, M. P. (2007). Mapping the functional connectivity of anterior cingulate cortex. Neuroimage, 37, 579–588.
Miller, M. I., Beg, M. F., Ceritoglu, C., & Stark, C. (2005). Increasing the power of functional maps of the medial temporal lobe by using large deformation diffeomorphic metric mapping. Proceedings of the National Academy of Sciences of the United States of America, 102, 9685–9690.
Molloy, E. K., Meyerand, M. E., & Birn, R. M. (2014). The influence of spatial resolution and smoothing on the detectability of resting-state and task fMRI. Neuroimage, 86, 221–230. https://doi.org/10.1016/j.neuroimage.2013.09.001.
Murphy, K., Bodurka, J., & Bandettini, P. A. (2007). How long to scan? The relationship between fMRI temporal signal to noise ratio and necessary scan duration. Neuroimage, 34(2), 565–574. https://doi.org/10.1016/j.neuroimage.2006.09.032.
Murphy, K., van Ginneken, B., Reinhardt, J. M., Kabus, S., Ding, K., Deng, X., et al. (2011). Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Transactions on Medical Imaging, 30, 1901–1920.
Peltier, S. J., & Noll, D. C. (2002). T2* dependence of low frequency functional connectivity. Neuroimage, 16, 985–992.
Peyrat, J.-M., Delingette, H., Sermesant, M., Xu, C., & Ayache, N. (2010). Registration of 4d cardiac ct sequences under trajectory constraints with multi-channel diffeomorphic demons. IEEE Transactions on Medical Imaging, 29, 1351–1368.
Poldrack, R. A., & Gorgolewski, K. J. (2014). Making big data open: data sharing in neuroimaging. Nature Neuroscience, 17(11), 1510–1517. https://doi.org/10.1038/nn.3818.
Poldrack, R. A., Baker, C. I., Durnez, J., Gorgolewski, K. J., Matthews, P. M., Munafo, M. R., et al. (2017). Scanning the horizon: towards transparent and reproducible neuroimaging research. Nature Reviews. Neuroscience, 18(2), 115–126. https://doi.org/10.1038/nrn.2016.167.
Purdon, P. L., & Weisskoff, R. M. (1998). Effect of temporal autocorrelation due to physiological noise and stimulus paradigm on voxel-level false-positive rates in fMRI. Human Brain Mapping, 6, 239–249.
Rueckert, D., Sonoda, L. I., Hayes, C., Hill, D. L., Leach, M. O., & Hawkes, D. J. (1999). Nonrigid registration using free-form deformations: application to breast mr images. IEEE Transactions on Medical Imaging, 18, 712–721.
Russell, J. F. (2013). If a job is worth doing, it is worth doing twice. Nature, 496, 7.
Schizophrenia Working Group of the Psychiatric Genomics, C. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511(7510), 421–427. https://doi.org/10.1038/nature13595.
Schmaal, L., Hibar, D. P., Samann, P. G., Hall, G. B., Baune, B. T., Jahanshad, N., et al. (2016). Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Molecular Psychiatry, 22(6), 900–909. https://doi.org/10.1038/mp.2016.60.
Shelton, D., Stetten, G., Aylward, S., Ibáñez, L., Cois, A., & Stewart, C. (2005). Teaching medical image analysis with the insight toolkit. Medical Image Analysis, 9, 605–611.
Smith, S. M., Jenkinson, M., Woolrich, M. W., Beckmann, C. F., Behrens, T. E. J., Johansen-Berg, H., Bannister, P. R., de Luca, M., Drobnjak, I., Flitney, D. E., Niazy, R. K., Saunders, J., Vickers, J., Zhang, Y., de Stefano, N., Brady, J. M., & Matthews, P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage, 23, S208–S219. https://doi.org/10.1016/j.neuroimage.2004.07.051.
Smith, S. M., Fox, P. T., Miller, K. L., Glahn, D. C., Fox, P. M., Mackay, C. E., Filippini, N., Watkins, K. E., Toro, R., Laird, A. R., & Beckmann, C. F. (2009). Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America, 106, 13040–13045.
Smith, S. M., Beckmann, C. F., Andersson, J., Auerbach, E. J., Bijsterbosch, J., Douaud, G., Duff, E., Feinberg, D. A., Griffanti, L., Harms, M. P., Kelly, M., Laumann, T., Miller, K. L., Moeller, S., Petersen, S., Power, J., Salimi-Khorshidi, G., Snyder, A. Z., Vu, A. T., Woolrich, M. W., Xu, J., Yacoub, E., Uğurbil, K., van Essen, D., Glasser, M. F., & WU-Minn HCP Consortium. (2013). Resting-state fMRI in the human connectome project. Neuroimage, 80, 144–168. https://doi.org/10.1016/j.neuroimage.2013.05.039.
Song, X., Zhang, Y., & Liu, Y. (2014). Frequency specificity of regional homogeneity in the resting-state human brain. PLoS One, 9(1), e86818.
Stanley, M. L., Moussa, M. N., Paolini, B. M., Lyday, R. G., Burdette, J. H., & Laurienti, P. J. (2013). Defining nodes in complex brain networks. Frontiers in Computational Neuroscience, 7, 169. https://doi.org/10.3389/fncom.2013.00169.
Stein, J. L., Medland, S. E., Vasquez, A. A., Hibar, D. P., Senstad, R. E., Winkler, A. M., et al. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44(5), 552–561. https://doi.org/10.1038/ng.2250.
Thompson, P. M., Stein, J. L., Medland, S. E., Hibar, D. P., Vasquez, A. A., Renteria, M. E., et al. (2014). The ENIGMA consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior, 8, 153–182. https://doi.org/10.1007/s11682-013-9269-5.
Triantafyllou, C., Hoge, R. D., Krueger, G., Wiggins, C. J., Potthast, A., Wiggins, G. C., & Wald, L. L. (2005). Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. Neuroimage, 26(1), 243–250. https://doi.org/10.1016/j.neuroimage.2005.01.007.
Triantafyllou, C., Hoge, R. D., & Wald, L. L. (2006). Effect of spatial smoothing on physiological noise in high-resolution fMRI. Neuroimage, 32(2), 551–557.
Turner, G. H., & Twieg, D. B. (2005). Study of temporal stationarity and spatial consistency of fMRI noise using independent component analysis. IEEE Transactions on Medical Imaging, 24(6), 712–718.
van Dalen, J. A., Vogel, W., Huisman, H. J., Oyen, W. J. G., Jager, G. J., & Karssemeijer, N. (2004). Accuracy of rigid CT-FDG-PET image registration of the liver. Physics in Medicine and Biology, 49, 5393–5405.
van Erp, T. G., Hibar, D. P., Rasmussen, J. M., Glahn, D. C., Pearlson, G. D., Andreassen, O. A., et al. (2016). Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Molecular Psychiatry, 21(4), 547–553. https://doi.org/10.1038/mp.2015.63.
Veraart, J., Fieremans, E., & Novikov, D. S. (2016a). Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Medicine, 76, 1582–1593. https://doi.org/10.1002/mrm.26059.
Veraart, J., Novikov, D. S., Christiaens, D., Ades-Aron, B., Sijbers, J., & Fieremans, E. (2016b). Denoising of diffusion MRI using random matrix theory. Neuroimage, 142, 394–406. https://doi.org/10.1016/j.neuroimage.2016.08.016.
Villain, N., Landeau, B., Groussard, M., Mevel, K., Fouquet, M., Dayan, J., Eustache, F., Desgranges, B., & Chételat, G. (2010). A simple way to improve anatomical mapping of functional brain imaging. Journal of Neuroimaging, 20(4), 324–333. https://doi.org/10.1111/j.1552-6569.2010.00470.x.
Weisskoff, R. M. (1996). Simple measurement of scanner stability for functional NMR imaging of activation in the brain. Magnetic Resonance in Medicine, 36(4), 643–645.
Welvaert, M., & Rosseel, Y. (2013). On the definition of signal-to-noise ratio and contrast-to-noise ratio for FMRI data. PLoS One, 8(11), e77089. https://doi.org/10.1371/journal.pone.0077089.
Woods, R. (1996). Correlation of brain structure and function. In A. Toga, J. Mazziotta (Ed.), Brain mapping: The methods (pp. 313–342). Academic Press.
Yoo, T. S., Ackerman, M. J., Lorensen, W. E., Schroeder, W., Chalana, V., Aylward, S., Metaxas, D., & Whitaker, R. (2002). Engineering and algorithm design for an image processing API: a technical report on ITK - the insight toolkit. Studies in Health Technology and Informatics, 85, 586–592.
Zhang, S., Li, X., Lv, J., Jiang, X., Guo, L., & Liu, T. (2016). Characterizing and differentiating task-based and resting state fMRI signals via two-stage sparse representations. Brain Imaging and Behavior, 10(1), 21–32. https://doi.org/10.1007/s11682-015-9359-7.
Zuo, X. N., Anderson, J. S., Bellec, P., Birn, R. M., Biswal, B. B., Blautzik, J., Breitner, J. C. S., Buckner, R. L., Calhoun, V. D., Castellanos, F. X., Chen, A., Chen, B., Chen, J., Chen, X., Colcombe, S. J., Courtney, W., Craddock, R. C., di Martino, A., Dong, H. M., Fu, X., Gong, Q., Gorgolewski, K. J., Han, Y., He, Y., He, Y., Ho, E., Holmes, A., Hou, X. H., Huckins, J., Jiang, T., Jiang, Y., Kelley, W., Kelly, C., King, M., LaConte, S. M., Lainhart, J. E., Lei, X., Li, H. J., Li, K., Li, K., Lin, Q., Liu, D., Liu, J., Liu, X., Liu, Y., Lu, G., Lu, J., Luna, B., Luo, J., Lurie, D., Mao, Y., Margulies, D. S., Mayer, A. R., Meindl, T., Meyerand, M. E., Nan, W., Nielsen, J. A., O’Connor, D., Paulsen, D., Prabhakaran, V., Qi, Z., Qiu, J., Shao, C., Shehzad, Z., Tang, W., Villringer, A., Wang, H., Wang, K., Wei, D., Wei, G. X., Weng, X. C., Wu, X., Xu, T., Yang, N., Yang, Z., Zang, Y. F., Zhang, L., Zhang, Q., Zhang, Z., Zhang, Z., Zhao, K., Zhen, Z., Zhou, Y., Zhu, X. T., & Milham, M. P. (2014). An open science resource for establishing reliability and reproducibility in functional connectomics. Scientific Data, 1, 140049. https://doi.org/10.1038/sdata.2014.49.
Funding
Support was received from NIH grants U54 EB020403, U01MH108148, 2R01EB015611, R01MH112180, R01DA027680, R01MH085646. MarbG and MuenG (FOR2107 study): Work was supported by the German Research Foundation (DFG), grant numbers FOR2107; KI 588/15–1 to TK, DA 1151/5–1 to UD, KO 4291/4–1 and KR 3822/5–1 to AK. JV is a Postdoctoral Fellow of the Research Foundation - Flanders (FWO; grant number 12S1615N).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest for any of the authors.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(DOCX 40 kb)
Rights and permissions
About this article
Cite this article
Adhikari, B.M., Jahanshad, N., Shukla, D. et al. A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: an ENIGMA rs-fMRI protocol. Brain Imaging and Behavior 13, 1453–1467 (2019). https://doi.org/10.1007/s11682-018-9941-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-018-9941-x