Abstract
Purpose
Careful assessment of the aortic root is paramount to select an appropriate prosthesis for transcatheter aortic valve implantation (TAVI). Relevant information about the aortic root anatomy, such as the aortic annulus diameter, can be extracted from pre-interventional CT. In this work, we investigate a neural network-based approach for segmenting the aortic root as a basis for obtaining these parameters.
Methods
To support valve prosthesis selection, geometric measures of the aortic root are extracted from the patient’s CT scan using a cascade of convolutional neural networks (CNNs). First, the image is reduced to the aortic root, valve, and left ventricular outflow tract (LVOT); within that subimage, the aortic valve and ascending aorta are segmented; and finally, the region around the aortic annulus. From the segmented annulus region, we infer the annulus orientation using principal component analysis (PCA). The area-derived diameter of the annulus is approximated based on the segmentation of the aortic root and LVOT and the plane orientation resulting from the PCA.
Results
The cascade of CNNs was trained using 90 expert-annotated contrast-enhanced CT scans routinely acquired for TAVI planning. Segmentation of the aorta and valve within the region of interest achieved an F1 score of 0.94 on the test set of 36 patients. The area-derived diameter within the annulus region was determined with a mean error below 2 mm between the automatic measurement and the diameter derived from annotations. The calculated diameters and resulting errors are comparable to published results of alternative approaches.
Conclusions
The cascaded neural network approach enabled the assessment of the aortic root with a relatively small training set. The processing time amounts to 30 s per patient, facilitating time-efficient, reproducible measurements. An extended training data set, including different levels of calcification or special cases (e.g., pre-implanted valves), could further improve this method’s applicability and robustness.
Similar content being viewed by others
Introduction
Transcatheter aortic valve implantation (TAVI) allows minimally invasive replacement of pathological aortic valves. However, purely image-based prosthetic valve selection is challenging.
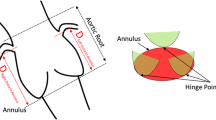
Figure 1 shows the complex anatomy of the aortic valve region. The implant needs to fit tight to the aortic wall and must not cover the coronary artery branches. Its orientation should be similar to the original valve. Therefore, it is essential to carefully analyze the aortic root before surgery to determine the best prosthesis and appropriate placement strategy. The assessment of the aortic valve annulus is crucial for prosthesis selection. Corresponding parameters such as the annulus diameter are highly sensitive to the orientation of the measurement plane. Several studies investigated the reliability and repeatability of interactive measurements: Meyer et al. [10] observed a difference in resulting prosthesis size between two software solutions in 18% of the considered patients. Schuhbaeck et al. [14] report mean inter-observer differences of \(0.4\pm 0.9\) mm with a maximum of 5.6 mm for the area-derived annulus diameter.
The blue bounding box contains the complete TAVI planning CT scan. The preprocessing for the aortic root detection crops this volume to the yellow region of \(256\times 256\times 384\) mm\(^3\) centered at a point 128 mm above the image center (in HEAD direction, according to the DICOM information). The subimage in the red bounding box contains the aortic root. The first task in this work is the automatic extraction of this subimage
We aim to support aortic root analysis with a fully automatic, deterministic and patient-specific approach, facilitating standardization and reduction of effort for TAVI planning.
We break down the task into a cascade of neural networks as suggested for related contexts [12]:
-
1.
Detection of a uniformly sized bounding box around the aortic valve;
-
2.
Segmentation of the aorta including aortic valve within this bounding box;
-
3.
Approximation of the area-derived annulus diameter based on the segmentation of the aortic valve and the annulus region.
We aim at developing an approach, applicable to a wide range of CT scanner models. We test our approach on expert-annotated routine clinical data acquired by different CT scanners and compare the suggested implant size based on the approximated annulus diameter with the implanted device size.
Related work
This section discusses the reviewed literature on segmentation of the aortic root region in contrast-enhanced CT images. Lalys et al. presented a comprehensive pipeline for TAVI analysis, using atlas registration and deformable 3D snakes [8]. Their approach requires the placement of a point in the aortic root region. Elattar et al. used fuzzy classification and normalized cuts to segment the aortic root after detecting the centerline [3]. They reported smooth aortic root surfaces, possibly affecting accuracy in subsequent measurements. Gao et al. proposed atlas-based segmentation followed by surface model fitting, with atlas images from a publicly available database [4]. Waechter et al. segmented the aortic root region with a mean-shape model [15]. Astudillo et al. described a CNN-based segmentation in manually defined annulus planes [2].
The approaches from Astudillo et al. and Lalys et al. require manual interaction, hindering implementation as a fully automatic and deterministic processing tool. The aortic root models of Elattar et al., Gao et al. and Waechter et al. are smoothed or based on a smooth model. The model’s deformation is limited and thus only yields an approximation of the patient-specific anatomy.
Given the reliable performance of CNN methods in recent years, our aim is a conceptually simple and fully automatic pipeline consisting of multiple hierarchical CNN steps.
Methods
We propose a neural network-based three-step segmentation approach for the anatomical structures of interest from CT images. The first step extracts the region of interest (ROI) around the aortic root and LVOT (see Fig. 2).
Each resulting subimage is resampled to an isotropic resolution of 0.6 mm. This is close to the original image resolution, which varies between 0.38 and 0.7 mm and ensures that processing with the given hardware is possible (see “Appendix”—hardware). In the second step, the aorta, including the aortic valve, is segmented within this ROI. This segmentation serves as additional input for estimating the annulus diameter. In the third step, the region around the aortic annulus is segmented, and the orientation of the annulus plane is determined using principal component analysis (PCA). Based on this information, we approximate the annulus diameter. Figure 3 visualizes the sequence of steps.
Data
Our data set comprises annotated 3D thorax CT scans of 126 patients from the German Heart Center Berlin and the Charité TAVI registry. We used 36 cases for testing. The remaining 90 cases were repeatedly split into 75 for training and 15 for validation in sixfold cross-validation.
Further, we collected a data set of 640 patients, who successfully underwent a TAVI procedure, to test the suitability of our model for device selection. A procedure was deemed successful when no complications were documented. Here, we compared the size of the implanted valve with our approximated annulus diameter. Patient statistics can be seen in Table 1.
All CTs of the training and sizing data set were acquired with Siemens scanners. In the test set, nine cases each were acquired with Siemens, Toshiba, Philips and GE scanners.
The image volumes have slice dimensions of 512 \(\times \) 512. The number of slices ranges from 273 to 1908, slice thickness 0.5–3 mm and spacing between slices 0.4–3 mm. The annotations shown in Fig. 4 were generated by domain experts using a custom MeVisLabFootnote 1-based software prototype to obtain:
-
a mask of the aortic lumen,
-
the centerline through the aorta and LVOT,
-
cross-sectional contours of the aorta and LVOT perpendicular to the centerline and
-
three markers, indicating the hinge points of the aortic valve cusps.
The aorta mask includes lumen and valve without LVOT, and the cross-sectional contours show the outer contour of the aortic root and LVOT.
Model training
For image processing, we considered the U-Net architecture, which has been successfully applied in several biomedical image segmentation challenges.Footnote 2 We trained a structurally identical CNN for each processing step [11]. The specific layer arrangement is illustrated in Fig. 5.
It consists of a contracting path and an expansive path. In our application, the contracting path has nine convolutional layers combined with batch normalization, ReLU activation, max pooling and dropout. The expansive path alternates convolutions with transposed, also called up-convolutions. As the last layer, a \(1\times 1\) convolution with sigmoid activation is used, resulting in a voxel-wise classification of the input image.
The neural network’s memory requirement depends on the number of filters, batch size and size of the input image. It has been shown that smaller batch sizes can improve accuracy and generalizability [5]. Thus, we selected a batch size of two for training. Given the size of the input images, the available hardware (see “Appendix”) allowed for a number of 18 filters in the first layer.
The Hounsfield scale generally provides comparable intensities based on the tissue density. The usage of contrast agent, as well as differences in dosage settings, can, however, result in intensity variations. We apply intensity normalization using mean and standard deviation (see Fig. 6, for example).
In each epoch, each training image is augmented by translation. The image is randomly shifted between zero to five voxels in each direction. Because the patient orientation in the scanner is standardized, we performed no rotational augmentation. Deformations were not applied to avoid implausible anatomies.
The Adam optimizer [6] is used with different loss functions to find the best approach for each step. We considered the binary cross-entropy, as well as its variation, the Focal Loss [9], as we assume that the enhanced focus on the misclassified examples is appropriate in our scenario with the strong class imbalance.
-
Binary Cross-Entropy
$$\begin{aligned} L_{bc}(y, {\hat{y}}) = {\left\{ \begin{array}{ll} -log({\hat{y}}), &{} \text {if } y = 1\\ -log(1-{\hat{y}}), &{} \text {otherwise}, \end{array}\right. } \end{aligned}$$ -
Focal Loss with \(\gamma = 2\)
$$\begin{aligned} L_{f}(y, {\hat{y}})&= -(1-p_t)^{\gamma }log(p_t), \text {with} \\ p_t&= {\left\{ \begin{array}{ll} {\hat{y}}, &{} \text {if } y = 1\\ 1-{\hat{y}}, &{} \text {otherwise}, \end{array}\right. } \end{aligned}$$
where y is the target mask (1 if the voxel belongs to the segmentation and 0 otherwise) and \({\hat{y}}\) the predicted probability for each of the n voxels to belong to the segmentation.
We also evaluated the Tversky Loss, which is similar to the F1 score, but enables steering the emphasis on false-positive and false-negative classifications via the parameter \(\beta \) [13], as well as the Focal Tversky Loss [1].
-
Tversky Loss with \(\beta \in [0.45, 0.50, 0.55,..., 0.95]\)
$$\begin{aligned} L_{t}(y, {\hat{y}})&= 1-T(y, {\hat{y}}), \text {with} \\ T(y, {\hat{y}})&= \frac{y{\hat{y}}}{y{\hat{y}} + \beta (1-y){\hat{y}} + (1-\beta )y(1-{\hat{y}})} \end{aligned}$$ -
Focal Tversky Loss with \(\beta \in [0.45, 0.50, 0.55,..., 0.95]\) and \(\gamma = 2\)
$$\begin{aligned} L_{ft}(y, {\hat{y}}) = (1-T(y, {\hat{y}}))^{1/\gamma } \end{aligned}$$
An individual model for each step is obtained by selecting the four loss function and parameter combinations with the best results on a validation set of 15 patients after training on 75 patients, then training six structurally identical models in sixfold cross-validation per each of the four selected loss functions and averaging their outputs. We use this bagging approach to exploit the benefits of ensemble methods, exemplified by the results for step 1.
Step 1: Aortic valve detection
In the first step, the image is cropped to a ROI around the aortic root and LVOT: The ROI detection is performed on a low-resolution (isotropic voxel size of 2 mm) subimage with an extent of \(256\times 256\times 384\) mm\(^3\) placed 128 mm above the image center (in the HEAD direction provided by the DICOM information) to enable fast memory-efficient processing. This image is padded if the upper border is outside the original image (see Fig. 2).
Training Based on the expert-defined aortic valve hinge points, an axis-aligned bounding box of \(80\times 80\times 80\) mm\(^3\) is placed in the image around the aortic root and LVOT, centered on the midpoint of the hinge points. This bounding box is used as a mask to learn the detection of the ROI.
The labels for the training of the ROI segmentation are generated based on the aortic valve hinge points via the calculation of an axis-aligned enclosing bounding box. The labels for the ensuing aorta segmentation in step 2 are based on the aortic lumen, while the region around the annulus plane is defined by the cross-sectional contours of the aortic root and LVOT
Segmentation postprocessing For each voxel in the input CT, the CNN returns a probability for this voxel to belong to the uniformly sized ROI. This is binarized with a threshold of 0.5, and a bounding box is drawn around all predicted voxels. This box is resized to the desired dimensions of \(80\times 80\times 80\) mm\(^3\) with the midpoint being the center of the predicted bounding box.
Step 2: Aorta segmentation
In the second step, a CNN is trained on the aorta segmentation within the ROI. The original images are resampled to an isotropic voxel size of 0.6 mm before being cropped to the ROI.
Training The label generation for this step is based on the aortic lumen within the ROI (see Fig. 7).
Segmentation postprocessing No postprocessing is required in this step.
Step 3: Aortic root analysis
In the third step, the region around the aortic annulus plane, defined by the three hinge points, is segmented to deduce the orientation and midpoint of the annulus plane and enable measurements of the annulus region. As an output of step 3, the aorta segmentation from step 2 is reduced to the valve cusps and combined with the segmentation of the annulus region from step 3 to achieve a segmentation comprising valve cusps, annulus and LVOT. This segmentation is analyzed in cross sections oriented parallel to the derived annulus plane, and the diameter over all cross sections is measured to determine an approximation of the annulus diameter.
Training Based on the expert-defined hinge point markers (Fig. 4), we estimate the orientation of the annulus plane. To train the CNN, we consider the mask of the aortic root and LVOT along a 10 mm margin in the direction of the plane normal, as visualized in Fig. 8. The margin of 10 mm was selected experimentally to achieve a mask that can be successfully trained while also yielding an ideal approximation of the annulus plane from PCA. With this 3D approach, we intend to make our analysis robust against the input uncertainty of the hinge points.
Segmentation postprocessing A PCA is applied to deduce the annulus plane from the predicted segmentation (see Fig. 9). PCA is a dimensionality reduction method, which reduces a data set to its uncorrelated components that maximize variance. By definition of our annulus plane region, its diameter parallel to the annulus plane should be larger than its height. According to the literature, an annulus diameter typically exceeds 17 mm [14] and the height of our segmentation mask is defined to be 10 mm. Thus, the two components with the highest variance describe the orientation of the annulus plane. The vector orthogonal to the two largest eigenvectors of the PCA is taken as an estimate of the plane’s normal vector. The center of gravity of the predicted segmentation estimates the midpoint of the annulus plane. This midpoint and normal vector allow for approximation of the annulus plane. In our training data set, the mean Euclidean distance between the midpoint of the three hinge points and the midpoint deduced from PCA is 1.5 mm and the mean angle difference between the resulting normal vectors is \(1.6^{\circ }\).
Principal component analysis applied to the segmentation mask (yellow): The first two principal components maximize the variance of the underlying data while being perpendicular to each other. A plane results from the normal vector orthogonal to the two principal components. The plane’s midpoint is defined by the center of gravity of the underlying data
However, the result is misleading if a segmented annulus region has a height comparable to its diameter (Fig. 10).
To ensure that the PCA detects the correct orientation, the predicted segmentation of the annulus region is masked with the contour of the segmentation from step 2 (see Fig. 11). Undesired segments are removed by only considering the largest connected component. The input of the PCA is thereby more accurately aligned along the annulus plane; however, the center of gravity is shifted into the valve. In our training data set, the mean Euclidean distance between the midpoint of the three hinge points and the midpoint deduced from this adjustment to the PCA input is 6.8 mm and the mean angle difference between the resulting normal vectors is \(1.7^\circ \). The midpoint and normal vector resulting from the masked segmentation of the annulus region allow for the approximation of the annulus plane.
Aortic Root Analysis The aortic valve segmentation from the previous step is combined with the segmentation of the annulus region and reduced to a height of 10 mm along the resulting plane to approximate the aortic root region including aortic valve leaflets and LVOT. The minimal diameter within this region is calculated using the slice-wise convex hull contours aligned along the deduced annulus plane and obtaining the area-derived diameter for each. We only consider minima above 17 mm to ensure that all three aortic leaflets are contained within the contour; an aortic annulus diameter below 17 mm is unlikely [14]. The reference diameter, used as ground truth, is obtained in the same manner from the annotated data: The aortic valve and LVOT segmentation masks are intersected parallel to the plane defined by the hinge points, and the reference diameter is calculated as minimum area-derived diameter in the resulting cross sections within a range of \(\pm 5\) mm along the plane normal.
In addition to the annotated data set, we apply our method to 640 CT scans of patients who underwent a successful TAVI. We compare the size of the implanted device to our suggestion based on our area-derived diameter, inspired by the sizing strategy from Schuhbaeck et al. [14]: a 23-mm valve for annulus diameters of 19.5–22.5 mm, a 26-mm valve for 22.5–26.5 mm, a 29-mm valve for 26.5–29.5 mm; we further suggested a 20-mm valve for annulus diameters below 19.5 mm and a 34-mm valve for greater than 29.5 mm.
Results
The following subsections show the results per step.
Step 1: Detection of the region of interest
In the detection of the ROI, a bounding box with a fixed size around the aortic root and LVOT centered around the hinge points shall be found by segmentation. The detected bounding box \(BB_\mathrm{unet}\) is compared to the reference bounding box \(BB_\mathrm{ref}\) using the intersection over ground truth (IoG):
The resulting bounding box around the aortic root and LVOT is used as input for the following steps. The IoG is not directly applied to the predicted segmentation but to the bounding box obtained after thresholding and reshaping, as explained in the “Methods” section.
Table 2 shows the results for the individual loss functions reduced to the best parameters for clarity. For the final model, we selected the Tversky Loss with \(\beta =0.80\) as well as the Focal Tversky Loss with \(\beta =0.65\), \(\beta =0.85\) and \(\beta =0.90\), because they provide the highest IoG on the validation set (highlighted in italics). Table 3 shows the IoG for the selected loss functions evaluated on all 90 training samples and the 36 test samples.
The final model resulting from the bagging of several models shows a higher IoG than any of the individual models in Table 2.
Step 2: Aorta segmentation
In the second step, the aorta, including the aortic valve within the obtained bounding box, is segmented. The resulting mask \(M_\mathrm{unet}\) is compared to the reference label \(M_\mathrm{ref}\) with the F1 score (Dice coefficient), which considers precision \(\frac{\left| M_\mathrm{unet} \cap M_\mathrm{ref} \right| }{\left| M_\mathrm{unet} \right| }\) and recall \(\frac{\left| M_\mathrm{unet} \cap M_\mathrm{ref} \right| }{\left| M_\mathrm{ref} \right| }\):
Table 4 shows the F1 score for the four loss functions reduced to the best parameters.
Table 5 shows the F1 score obtained from the selected loss functions, Tversky Loss with \(\beta =0.55\) and Focal Tversky Loss with \(\beta =0.55\), \(\beta =0.65\) and \(\beta =0.70\), as well as the final model, which is again the averaged output of the models with these four loss functions.
Step 3: Aortic root analysis
The aortic root is found by considering the segmented aortic valve and LVOT in cross sections reformatted to the orientation of the detected valve plane. The minimal area-derived diameter within the aortic root is compared to the minimal area-derived diameter obtained from the annotated data set. Table 6 shows the mean error, which was used for the selection of the optimal loss functions, the Tversky Loss with \(\beta =0.55\), \(\beta =0.85\) and \(\beta =0.90\) and the Focal Tversky Loss with \(\beta =0.80\).
Table 7 shows the results for the automatically obtained diameters from the final model, the diameters deduced from the annotations and the respective error. Table 8 compares the test results of the different CT scanner manufacturers. No significant difference between the errors for individual manufacturers can be observed in our test data set (ANOVA, \(p =0.78\)).
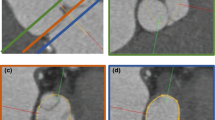
Possible error sources: a difference between the annotations (green) and the model (red) in detected plane orientation (lines), aorta and valve segmentation (silhouettes) and segmentation around the annulus plane (solid); in this example, this leads to a difference in the diameter obtained from the model and the annotations of 5.91 mm
Table 9 shows the results for implant size selection. We suggested the correct implant size in 81% of the training cases, 61% of the test cases and 64% of the device sizing cases.
On average, the processing time for one patient from the input of the original CT to the measurement of the minimal diameter amounts to 30 s, given the available hardware (see “Appendix”).
Discussion and conclusion
This paper aims to test the suitability of a neural network-based approach to support aortic root analysis for TAVI planning. Cascaded neural networks are applied for three processing steps: detection of the ROI around the aortic root and LVOT, segmentation of the aorta, including the aortic valve, and segmentation of the region around the aortic annulus. The inferred minimal area-derived diameter within the annulus region is compared to the diameter from expert annotations. The aortic root is measured in cross sections, whose orientation corresponds to the annulus plane. The annulus plane is approximated by applying a PCA to the segmentation of the annulus region, masked with the outer edge of the aortic valve segmentation.
On average, our calculated minimal area-derived diameter within the annulus region deviates less than 2 mm from the minimal diameter from expert annotations. The calculated diameters are in a range comparable to similar studies [3, 8, 15]. The implant size could be inferred in 81% of the cases for the training set, 62% for the test set and 64% for the device sizing set. No significant difference between different CT scanners could be observed.
The accuracy of the model’s predictions depends on the annotations of the data set. For the present study, the training set comprises a relatively small number of 90 cases and 36 cases are used for the evaluation of the model. Further improvement and evaluation with a larger data set are advisable.
Future work is also required to optimize the detection of the aortic annulus plane. We currently only consider the plane orientation, while its exact position is estimated by the minimal area-derived diameter in the annulus region.
The measured diameters are highly dependent on several factors, such as the orientation of the detected annulus plane. Kütting et. al report that plane tilting leads to a different choice in valve prosthesis size in 33% of cases for \(5^{\circ }\) of tilt and 66% of cases for \(10^{\circ }\) [7]. Figure 12 shows an example of possible error sources—a difference in detected plane orientation, aortic valve segmentation and segmentation around the annulus plane. In this example, all error sources are present.
The optimal valve size does not only depend on the annulus diameter but also other factors, such as the coronary distances, calcifications or aortic dilation. Although the strategy for prosthesis sizing in the clinical setting currently also concentrates on the annulus measurement, we suggest consideration of the more complex three-dimensional aortic root anatomy by, for example, regarding several planes throughout the aortic root along the centerline. This would allow for a thorough analysis of the aortic root contour and diminish the effect of measurement of a single tilted annulus plane. For such an extensive aortic root analysis, an automatic approach would be highly valuable for time efficiency and reproducibility of results.
The current results suggest neural network-based aortic root analysis as a promising approach to support fully automatic, time-efficient and reproducible aortic root assessment for TAVI device sizing and selection, which can be improved through enhanced training data on the one hand and more sophisticated consideration of the aortic root anatomy on the other hand.
Data availability
Not publicly available.
Notes
The ISBI 2015: https://biomedicalimaging.org/2015/, where the U-Net won two challenges: https://lmb.informatik.uni-freiburg.de/people/ronneber/isbi2015/.
References
Abraham N, Khan NM (2019) A novel focal Tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019), pp 683–687. https://doi.org/10.1109/ISBI.2019.8759329
Astudillo P, Mortier P, Bosmans J, De Backer O, de Jaegere P, De Beule M, Dambre J (2019) Enabling automated device size selection for transcatheter aortic valve implantation. J Interv Cardiol 2019:1–7. https://doi.org/10.1155/2019/3591314
Elattar M, Wiegerinck EM, van Kesteren F, Dubois L, Planken N, Vanhavel E, Baan J, Marquering H (2016) Automatic aortic root landmark detection in CTA images for preprocedural planning of transcatheter aortic valve implantation. Int J Cardiovasc Imaging 32:501–511
Gao X, Kitslaar PH, Reiber JHC (2016) Automatic aortic root segmentation in CTA whole-body dataset. In: Proceedings of SPIE 9785, medical imaging 2016: computer-aided diagnosis, p 8
Kandel I, Castelli M (2020) The effect of batch size on the generalizability of the convolutional neural networks on a histopathology dataset. ICT Express 6(4):312–315. https://doi.org/10.1016/j.icte.2020.04.010
Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. CoRR arXiv:1412.6980
Kütting M, Sedaghat A, Tapia AW, Roggenkamp J, Werner N, Schmitz-Rode T, Steinseifer U (2013) Influence of the measurement plane on aortic annulus indices: structural and clinical implications. Cardiovasc Eng Technol 4:513–519. https://doi.org/10.1007/s13239-013-0154-6
Lalys F, Esneault S, Castro M, Royer L, Haigron P, Auffret V, Tomasi J (2019) Automatic aortic root segmentation and anatomical landmarks detection for TAVI procedure planning. Minim Invasive Ther Allied Technol 28(3):157–164. https://doi.org/10.1080/13645706.2018.1488734
Lin TY, Goyal P, Girshick R, He K, Dollár P (2020) Focal loss for dense object detection. IEEE Trans Pattern Anal Mach Intell 42(2):318–327. https://doi.org/10.1109/TPAMI.2018.2858826
Meyer A, Kofler M, Montagner M, Unbehaun A, Sündermann S, Buz S, Klein C, Stamm C, Solowjowa N, Emmert MY, Falk V, Kempfert J (2020) Reliability and influence on decision making of fully-automated vs. semi-automated software packages for procedural planning in tavi. Sci Rep 10:10746. https://doi.org/10.1038/s41598-020-67111-5
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds) Medical image computing and computer-assisted intervention—MICCAI 2015. Springer International Publishing, Cham, pp 234–241
Roth HR, Oda H, Zhou X, Shimizu N, Yang Y, Hayashi Y, Oda M, Fujiwara M, Misawa K, Mori K (2018) An application of cascaded 3d fully convolutional networks for medical image segmentation. Comput Med Imaging Graph 66:90–99. https://doi.org/10.1016/j.compmedimag.2018.03.001
Salehi SSM, Erdogmus D, Gholipour A (2017) Tversky loss function for image segmentation using 3d fully convolutional deep networks. In: Wang Q, Shi Y, Suk HI, Suzuki K (eds) Machine learning in medical imaging. Springer International Publishing, Cham, pp 379-387
Schuhbaeck A, Achenbach S, Pflederer T, Marwan M, Schmid J, Nef H, Rixe J, Hecker F, Schneider C, Lell M, Uder M, Arnold M (2014) Reproducibility of aortic annulus measurements by computed tomography. Eur Radiol 24:1887–1888. https://doi.org/10.1007/s00330-014-3199-5
Waechter I, Kneser R, Korosoglou G, Peters J, Bakker NH, Boomen Rvd, Weese J (2010). Patient specific models for planning and guidance of minimally invasive aortic valve implantation, pp 526–533. https://doi.org/10.1007/978-3-642-15705-9_64
Open Access
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant agreement \(\hbox {N}^{\circ }\) 101017578 and the BMBF Project TRANSNAV (13GW0372D).
Author information
Authors and Affiliations
Contributions
NK, AM, AH, LT and MH conceived the study and designed the methods. NK, AM, MK, AS, LT, MH, IW, MP and AH obtained and prepared the data. Falk and Kempfert gave medical and scientific advice. The first draft of the manuscript was written by NK, LT and AH. All authors commented on previous versions of the manuscript, read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethics approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Need for informed consent was waived due to the retrospective nature of the study.
Code availability
Not publicly available.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Hardware and software specifications
A Linux server with the following hardware specifications was used for model development:
-
2 Intel Xeon Gold 5215 @ 1.7 GHz,
-
4 nVidia Titan V 12 GB HBM2 (GV100, SM7.0).
-
96 GB RAM
Depending on the regarded step, the average training time of one CNN ranged between 20 minutes and one hour.
We used Keras 2.3.1 with the TensorFlow backend (tensorflow-gpu 2.1.0).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krüger, N., Meyer, A., Tautz, L. et al. Cascaded neural network-based CT image processing for aortic root analysis. Int J CARS 17, 507–519 (2022). https://doi.org/10.1007/s11548-021-02554-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02554-3