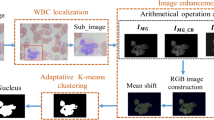
Abstract
The precise segmentation of white blood cells (WBCs) within blood smear images is a significant challenge with implications for both medical research and image processing. Of particular importance is the often neglected task of accurately segmenting WBC nuclei, an aspect that currently lacks dedicated methodologies. This paper introduces a straightforward and efficient method designed to fill this critical gap, providing an effective solution for the efficient segmentation of WBC nuclei. In blood smear imagery, the distinctive coloration of WBCs contrasts with the hues of other blood components. The inherent obscurity of WBCs prompts their segmentation by isolating pixels with minimal intensities. To streamline this process, our proposed method employs the Laplacian pyramid technique to decorrelate pixels in blood smear images, thereby amplifying the contrast. Subsequently, the intensities of pixels constituting blood cells, encompassing WBCs and the background, are modeled using three Gaussian random variables. Capitalizing on this feature, we implement the Gaussian mixture model (GMM) clustering method to determine the optimal threshold value, facilitating a highly precise segmentation of WBC nuclei. The proposed method demonstrates the capability to process images containing a single WBC as well as effectively functioning with images containing multiple cells of this type. Evaluation of the method on the ALL-IDB, ALL-IDB2, CellaVision, and JTSC datasets yielded accuracy values of 0.9802, 0.9725, 0.9772, and 0.9730, respectively. Comparative analysis with state-of-the-art methods revealed a notably comparable performance, underscoring the effectiveness of the proposed approach. The method presented in this article is highly competitive for segmenting the nuclei of WBCs compared to state-of-the-art methods. The three main advantages of our method are its ability to process images containing one or more WBCs, the automatic calculation of threshold values for each processed image, eliminating the need for manual parameter adjustments. Lastly, the method is efficient, as its algorithmic complexity is approximately \(\varvec{\mathcal {O}(nm)}\).
Graphical abstract











Similar content being viewed by others
References
Liu H, Cao H, Song E (2019) Bone marrow cells detection: a technique for the microscopic image analysis. J Med Syst 43:82. https://doi.org/10.1007/s10916-019-1185-9
Mishra S, Majhi B, Sa PK (2019) Texture feature based classification on microscopic blood smear for acute lymphoblastic leukemia detection. Biomed Signal Process Control 47:303–311. https://doi.org/10.1016/j.bspc.2018.08.012
Parente J (2019) Diagnosis for white blood cell abnormalities: leukocytes and leukopenia. Physician Assist Clin 4(3):625–635. https://doi.org/10.1016.j.cpha.2019.02.010
Al-Dulaimi K, Banks J, Nugyen K, Al-Sabaawi A, Tomeo-Reyes I, Chandran V (2021) Segmentation of white blood cell, nucleus and cytoplasm in digital haematology microscope images: a review-challenges, current and future potential techniques. IEEE Rev Biomed Eng 14:290–306. https://doi.org/10.1109/RBME.2020.3004639
Shahzad M, Umar AI, Khan MA, Shirazi SH, Khan Z, Yousaf W (2020) Robust method for semantic segmentation of whole-slide blood cell microscopic images. Comput Math Methods Med 2020:4015323
Li H, Zhao X, Su A, Zhang H, Liu J, Gu G (2020) Color space transformation and multi-class weighted loss for adhesive white blood cell segmentation. IEEE Access 8:24808–24818. https://doi.org/10.1109/ACCESS.2020.2970485
Li D, Yin S, Lei Y, Qian J, Zhao C, Zhang L (2023) Segmentation of white blood cells based on CBAM-DC-UNet. IEEE Access 11:1074–1082. https://doi.org/10.1109/ACCESS.2022.3233078
Lu Y, Qin X, Fan H, Lai T, Li Z (2021) WBC-Net: a white blood cell segmentation network based on UNet++ and ResNet. Appl Soft Comput 101:107006. https://doi.org/10.1016/j.asoc.2020.107006
Reena MR, Ameer PM (2021) Segmentation of leukocyte by semantic segmentation model: a deep learning approach. Biomed Signal Process Control 65:102385
Devi TG, Patil N, Rai S, Sarah CP (2023) Segmentation and classification of white blood cancer cells from bone marrow microscopic images using duplet-convolutional neural network design. Multimed Tools Appl. https://doi.org/10.1007/s11042-023-14899-9
Huynh HT, Dat VVT, Anh HB (2021) White blood cell segmentation and classification using deep learning coupled with image processing technique. In: Dang TK, Küng J, Chung TM, Takizawa M (eds) Future data and security engineering. Big data, security and privacy, smart city and industry 4.0 applications. Springer, Singapore, pp 399–410
Kumar PS, Vasuki S (2017) Automated diagnosis of acute lymphocytic leukemia and acute myeloid leukemia using multi-SV. J Biomed Imaging Bioeng 1(1):20–24
Hegde RB, Prasad K, Hebbar H, Singh BMK (2019) Development of a robust algorithm for detection of nuclei of white blood cells in peripheral blood smear images. Multimed Tools Appl 78:17879–17898
Mohd Safuan SN, Md Tomari MR, Wan Zakaria WN (2018) White blood cell (WBC) counting analysis in blood smear images using various color segmentation methods. Measurement 116:543–555. https://doi.org/10.1016/j.measurement.2017.11.002
Banik PP, Saha R, Kim K-D (2020) An automatic nucleus segmentation and CNN model based classification method of white blood cell. Expert Syst Appl 149:113211. https://doi.org/10.1016/j.eswa.2020.113211
Sudha K, Geetha P (2020) A novel approach for segmentation and counting of overlapped leukocytes in microscopic blood images. Biocybern Biomed Eng 40(2):639–648. https://doi.org/10.1016/j.bbe.2020.02.005
Bouchet A, Montes S, Ballarin V, Díaz I (2020) Intuitionistic fuzzy set and fuzzy mathematical morphology applied to color leukocytes segmentation. SIViP 14:557–564
Liu Y, Cao F, Zhao J, Chu J (2017) Segmentation of white blood cells image using adaptive location and iteration. IEEE J Biomed Health Inform 21(6):1644–1655. https://doi.org/10.1109/JBHI.2016.2623421
Makem M, Tiedeu A, Kom G, Kamdeu Nkandeu YP (2022) A robust algorithm for white blood cell nuclei segmentation. Multimed Tools Appl 81:17849–17874
Akram N, Adnan S, Asif M, Imran SMA, Yasir MN, Naqvi RA, Hussain D (2022) Exploiting the multiscale information fusion capabilities for aiding the leukemia diagnosis through white blood cells segmentation. IEEE Access 10:48747–48760. https://doi.org/10.1109/ACCESS.2022.3171916
Shahin AI, Guo Y, Amin KM, Sharawi AA (2018) A novel white blood cells segmentation algorithm based on adaptive neutrosophic similarity score. Health Inf Sci Syst 6(1):1–12. https://doi.org/10.1007/s13755-017-0038-5
Cao F, Liu Y, Huang Z, Chu J, Zhao J (2019) Effective segmentations in white blood cell images using \(\epsilon \)-SVR-based detection method. Neural Comput Appl 31:6767–6780. https://doi.org/10.1007/s00521-018-3480-7
Settouti N, Saidi M, Bechar MEA, Daho MEH, Chikh MA (2020) An instance and variable selection approach in pixel-based classification for automatic white blood cells segmentation. Pattern Anal Applic 23:1709–1726. https://doi.org/10.1007/s10044-020-00873-w
Abdurrazzaq A, Junoh AK, Yahya Z, Mohd I (2021) New white blood cell detection technique by using singular value decomposition concept. Multimed Tools Appl 80:4627–4638. https://doi.org/10.1007/s11042-020-09946-8
Sayed GI, Solyman M (2019) A novel chaotic optimal foraging algorithm for unconstrained and constrained problems and its application in white blood cell segmentation. Neural Comput & Applic 31:7633–7664. https://doi.org/10.1007/s00521-018-3597-8
Mahanta LB, Bora K, Kalita SJ, Yogi P (2019) Automated counting of platelets and white blood cells from blood smear images. In: Deka B, Maji P, Mitra S, Bhattacharyya DK, Bora PK, Pal SK (eds) Pattern recognition and machine intelligence. Springer, Cham, pp 13–20
Devi TG, Patil N, Rai S, Philipose CS (2022) Survey of leukemia cancer cell detection using image processing. In: Raman B, Murala S, Chowdhury A, Dhall A, Goyal P (eds) Computer vision and image processing. Springer, Cham, pp 468–488
Burt P, Adelson E (1983) The Laplacian pyramid as a compact image code. IEEE Trans Commun COM–31(4):532–540
Gonzalez-Ruiz V, Garcia-Ortiz JP, Fernandez-Fernandez MR, Fernandez JJ (2022) Optical flow driven interpolation for isotropic FIB-SEM reconstructions. Comput Methods Prog Biomed 221:106856. https://doi.org/10.1016/j.cmpb.2022.106856
Andrade AR, Vogado LHS, de MS Veras R, Silva RRV, Araujo FHD, Medeiros FNS (2019) Recent computational methods for white blood cell nuclei segmentation: a comparative study. Comput Methods Prog Biomed 173:1–14
Prinyakupt J, Pluempitiwiriyawej C (2015) Segmentation of white blood cells and comparison of cell morphology by linear and Naïve Bayes classifiers. Biomed Eng Online 14:63
Makem M, Tiedeu A (2020) An efficient algorithm for detection of white blood cell nuclei using adaptive three stage PCA-based fusion. Inform Med Unlocked 20:100416
Zhong Z, Wang T, Zeng K, Zhou X, Li Z (2019) White blood cell segmentation via sparsity and geometry constraints. IEEE Access 7:167593–167604. https://doi.org/10.1109/ACCESS.2019.2954457
Stark BA (2017) Studying moments of the central limit theorem. Math Enthus 14(1):53–76. https://doi.org/10.54870/1551-3440.1388
Garcia-Lamont F, Cervantes J, Lopez A, Rodriguez L (2018) Segmentation of images by color features: a survey. Neurocomputing 292:1–27. https://doi.org/10.1016/j.neucom.2018.01.091
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Garcia-Lamont, F., Lopez-Chau, A., Cervantes, J. et al. Nucleus segmentation of white blood cells in blood smear images by modeling the pixels’ intensities as a set of three Gaussian distributions. Med Biol Eng Comput (2024). https://doi.org/10.1007/s11517-024-03065-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11517-024-03065-4