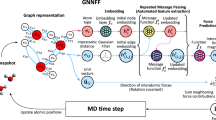
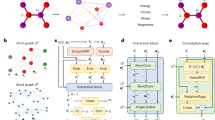
Abstract
Molecular dynamics is a powerful simulation tool to explore material properties. Most realistic material systems are too large to be simulated using first-principles molecular dynamics. Classical molecular dynamics has a lower computational cost but requires accurate force fields to achieve chemical accuracy. In this work, we develop a symmetry-adapted graph neural network framework called the molecular dynamics graph neural network (MDGNN) to construct force fields automatically for molecular dynamics simulations for both molecules and crystals. This architecture consistently preserves translation, rotation, and permutation invariance in the simulations. We also propose a new feature engineering method that includes high-order terms of interatomic distances and demonstrate that the MDGNN accurately reproduces the results of both classical and first-principles molecular dynamics. In addition, we demonstrate that force fields constructed by the proposed model have good transferability. The MDGNN is thus an efficient and promising option for performing molecular dynamics simulations of large-scale systems with high accuracy.
Similar content being viewed by others
References
R. Car, and M. Parrinello, Phys. Rev. Lett. 55, 2471 (1985).
K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov, and A. D. Mackerell Jr., J. Comput. Chem. 31, 671 (2009).
W. L. Jorgensen, D. S. Maxwell, and J. Tirado-Rives, J. Am. Chem. Soc. 118, 11225 (1996).
J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman, and D. A. Case, J. Comput. Chem. 25, 1157 (2004).
K. Hornik, M. Stinchcombe, and H. White, Neural Netw. 2, 359 (1989).
K. Hornik, Neural Netw. 4, 251 (1991).
J. Behler, and M. Parrinello, Phys. Rev. Lett. 98, 146401 (2007).
S. Chmiela, A. Tkatchenko, H. E. Sauceda, I. Poltavsky, K. T. Schütt, and K. R. Müller, Sci. Adv. 3, e1603015 (2017), arXiv: 1611.04678.
K. T. Schütt, F. Arbabzadah, S. Chmiela, K. R. Müller, and A. Tkatchenko, Nat. Commun. 8, 13890 (2017), arXiv: 1609.08259.
J. Han, L. Zhang, R. Car, and W. E, arXiv: 1707.01478.
L. Zhang, J. Han, H. Wang, R. Car, and W. E, Phys. Rev. Lett. 120, 143001 (2018), arXiv: 1707.09571.
H. Wang, L. Zhang, J. Han, and W. E, Comput. Phys. Commun. 228, 178 (2018), arXiv: 1712.03641.
S. Chmiela, H. E. Sauceda, K.-R. Müller, and A. Tkatchenko, Nat. Commun. 9, 3887 (2018).
L. Zhang, J. Han, H. Wang, W. Saidi, R. Car, W. E, in End-to-end symmetry preserving inter-atomic potential energy model for finite and extended systems: Proceedings of the 32nd International Conference on Neural Information Processing Systems (Montréal, Canada, 2018), p. 4436.
F. Scarselli, M. Gori, M. A. C. Tsoi, M. Hagenbuchner, and G. Monfardini, IEEE Trans. Neural Netw. 20, 61 (2009).
W. L. Hamilton, R. Ying, and J. Leskovec, arXiv: 1709.05584.
J. Bruna, W. Zaremba, A. Szlam, and Y. LeCun, arXiv: 1312.6203.
J. Zhou, G. Cui, Z. Zhang, C. Yang, Z. Liu, L. Wang, C. Li, and M. Sun, arXiv: 1812.08434.
Z. Wu, S. Pan, F. Chen, G. Long, C. Zhang, and S. Y. Philip, IEEE Trans. Neural Netw. Learn Syst. 32, 4 (2020).
Z. Zhang, P. Cui, and W. Zhu, IEEE Trans. Knowl. Data Eng. 1, 1 (2020).
K. Schütt, P.-J. Kindermans, H. E. S. Felix, S. Chmiela, A. Tkatchenko, and K.-R. Müller, in SchNet: A continuous-filter convolutional neural network for modeling quantum interactions: Proceeding of the 31st Conference on Neural Information Processing Systems (NIPS 2017) (Long Beach Convention Center, Long Beach, 2017), pp. 991–1001.
K. T. Schütt, H. E. Sauceda, P. J. Kindermans, A. Tkatchenko, and K. R. Müller, J. Chem. Phys. 148, 241722 (2018).
B. Anderson, T. S. Hy, and R. Kondor, arXiv: 1906.04015.
J. Klicpera, J. Groß, and S. Günnemann, in Directional message passing for molecular graphs: International Conference on Learning Representations (Addis Ababa, Ethiopia, 2019).
S. Takamoto, S. Izumi, and J. Li, arXiv: 1912.01398.
P. Frasconi, M. Gori, and A. Sperduti, IEEE Trans. Neural Netw. 9, 768 (1998).
A. Sperduti, and A. Starita, IEEE Trans. Neural Netw. 8, 714 (1997).
M. Hagenbuchner, A. Sperduti, and A. A. C. Tsoi, IEEE Trans. Neural Netw. 14, 491 (2003).
Q. Li, Z. Han, and X.-M. Wu, in Deeper insights into graph convolutional networks for semi-supervised learning: Proceeding of the Thirty-Second AAAI Conference on Artificial Intelligence (New Orleans, Riverside, 2018).
J. Gilmer, S. S. Schoenholz, P. F. Riley, O. Vinyals, and G. E. Dahl, in Neural message passing for quantum chemistry: Proceedings of the 34th International Conference on Machine Learning-Volume 70 (PMLR, 2017), pp. 1263–1272.
T. N. Kipf, and M. Welling, arXiv: 1609.02907.
Z. Ying, J. You, C. Morris, X. Ren, W. Hamilton, and J. Leskovec, in Hierarchical graph representation learning with differentiable pooling: Proceedings of the 32nd International Conference on Neural Information Processing Systems (Montréal, Canada, 2018), pp. 4800–4810.
M. Zhang, Z. Cui, M. Neumann, and Y. Chen, in An end-to-end deep learning architecture for graph classification: Proceeding of the Thirty-Second AAAI Conference on Artificial Intelligence (New Orleans, Riverside, 2018).
Y. Gal, and Z. Ghahramani, in A theoretically grounded application of dropout in recurrent neural networks: Advances in Neural Information Processing Systems 29 (NIPS 2016) (Barcelona, 2016), pp. 1019–1027.
K. Xu, C. Li, Y. Tian, T. Sonobe, K.-i. Kawarabayashi, and S. Jegelka, arXiv: 1806.03536.
K. He, X. Zhang, S. Ren, and J. Sun, in Deep residual learning for image recognition: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Las Vegas, 2016), pp. 770–778.
A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga, A. Desmaison, A. Kopf, E. Yang, Z. DeVito, M. Raison, A. Tejani, S. Chilamkurthy, B. Steiner, L. Fang, J. Bai, and S. Chintala, in Pytorch: An imperative style, high-performance deep learning library: Advances in Neural Information Processing Systems 32 (Montréal, Canada, 2019), pp. 8024–8035.
M. Fey, and J. E. Lenssen, in Fast graph representation learning with PyTorch Geometric: ICLR Workshop on Representation Learning on Graphs and Manifolds (New Orleans, 2019).
F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duchesnay, J. Mach. Learn Res. 12, 2825 (2011).
K. Xu, W. Hu, J. Leskovec, and S. Jegelka, arXiv: 1810.00826.
G. Li, M. Muller, A. Thabet, and B. Ghanem, in DeepGCNs: Can GCNs go as deep as CNNs? Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (Long Beach, 2019), pp. 9267–9276.
R. Hahnloser, R. Sarpeshkar, M. A. Mahowald, R. J. Douglas, and H. S. Seung, Nature 405, 947 (2000).
R. Hahnloser, and H. S. Seung, in Permitted and forbidden sets in symmetric threshold-linear networks: Conference on Neural Information Processing Systems (NeurIPS) (Denver, 2001), pp. 217–223.
X. Glorot, A. Bordes, and Y. Bengio, in Deep sparse rectifier neural networks: Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics (PMLR, 2011), pp. 315–323.
D. P. Kingma, and J. Ba, arXiv: 1412.6980.
G. Kresse, and J. Hafner, Phys. Rev. B 47, 558 (1993).
G. Kresse, and J. Hafner, Phys. Rev. B 49, 14251 (1994).
G. Kresse, and J. Furthmüller, Comput. Mater. Sci. 6, 15 (1996).
G. Kresse, and J. Furthmüller, Phys. Rev. B 54, 11169 (1996).
P. E. Blöchl, Phys. Rev. B 50, 17953 (1994).
G. Kresse, and D. Joubert, Phys. Rev. B 59, 1758 (1999).
J. P. Perdew, K. Burke, and M. Ernzerhof, Phys. Rev. Lett. 77, 3865 (1996).
J. P. Perdew, K. Burke, and M. Ernzerhof, Phys. Rev. Lett. 78, 1396 (1997).
C. A. Becker, F. Tavazza, Z. T. Trautt, and R. A. Buarque de Macedo, Curr. Opin. Solid State Mater. Sci. 17, 277 (2013).
L. M. Hale, Z. T. Trautt, and C. A. Becker, Model. Simul. Mater. Sci. Eng. 26, 055003 (2018).
A. Hjorth Larsen, J. Jörgen Mortensen, J. Blomqvist, I. E. Castelli, R. Christensen, M. Dułak, J. Friis, M. N. Groves, B. Hammer, C. Hargus, E. D. Hermes, P. C. Jennings, P. Bjerre Jensen, J. Kermode, J. R. Kitchin, E. Leonhard Kolsbjerg, J. Kubal, K. Kaasbjerg, S. Lysgaard, J. Bergmann Maronsson, T. Maxson, T. Olsen, L. Pastewka, A. Peterson, C. Rostgaard, J. Schiøtz, O. Schütt, M. Strange, K. S. Thygesen, T. Vegge, L. Vilhelmsen, M. Walter, Z. Zeng, and K. W. Jacobsen, J. Phys.-Condens. Matter 29, 273002 (2017).
S. R. Bahn, and K. W. Jacobsen, Comput. Sci. Eng. 4, 56 (2002).
F. Apostol, and Y. Mishin, Phys. Rev. B 83, 054116 (2011).
M. S. Daw, and M. I. Baskes, Phys. Rev. Lett. 50, 1285 (1983).
M. S. Daw, and M. I. Baskes, Phys. Rev. B 29, 6443 (1984).
X. Y. Liu, C. L. Liu, and L. J. Borucki, Acta Mater. 47, 3227 (1999).
N. Watters, D. Zoran, T. Weber, P. Battaglia, R. Pascanu, and A. Tacchetti, in Visual interaction networks: Learning a physics simulator from video: Advances in Neural Information Processing Systems 30 (NIPS 2017) (Long Beach, 2017), pp. 4539–4547.
C. R. Qi, H. Su, K. Mo, and L. J. Guibas, in Pointnet: Deep learning on point sets for 3D classification and segmentation: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Hawaii, 2017), pp. 652–660.
C. R. Qi, L. Yi, H. Su, and L. J. Guibas, in PointNet++: Deep Hierarchical Feature Learning on Point Sets in a Metric Space: Advances in Neural Information Processing Systems 30 (NIPS 2017) (Long Beach, 2017), pp. 5099–5108.
T. Xie, and J. C. Grossman, Phys. Rev. Lett. 120, 145301 (2018).
Author information
Authors and Affiliations
Corresponding authors
Additional information
This work was supported by the Basic Science Center Project of National Natural Science Foundation of China (Grant No. 51788104), the Ministry of Science and Technology of China (Grant Nos. 2016YFA0301001, and 2017YFB0701502), and the Beijing Advanced Innovation Center for Materials Genome Engineering.
Rights and permissions
About this article
Cite this article
Wang, Z., Wang, C., Zhao, S. et al. Symmetry-adapted graph neural networks for constructing molecular dynamics force fields. Sci. China Phys. Mech. Astron. 64, 117211 (2021). https://doi.org/10.1007/s11433-021-1739-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11433-021-1739-4