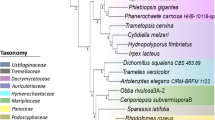
Abstract
The prospection of new degrading enzymes of the plant cell wall has been the subject of many studies and is fundamental for industries, due to the great biotechnological importance of achieving a more efficient depolymerization conversion from plant polysaccharides to fermentable sugars, which are useful not only for biofuel production but also for various bioproducts. Thus, we explored the shotgun metagenome data of a bacterial community (CB10) isolated from sugarcane bagasse and recovered three metagenome-assembled genomes (MAGs). The genomic distance analyses, along with phylogenetic analysis, revealed the presence of a putative novel Chitinophaga species, a Pandoraea nosoerga, and Labrys sp. isolate. The isolation process for each one of these bacterial lineages from the community was carried out in order to relate them with the MAGs. The recovered draft genomes have reasonable completeness (72.67–100%) and contamination (0.26–2.66%) considering the respective marker lineage for Chitinophaga (Bacteroidetes), Pandoraea (Burkholderiales), and Labrys (Rhizobiales). The in-vitro assay detected cellulolytic activity (endoglucanases) only for the isolate Chitinophaga, and its genome analysis revealed 319 CAZymes, of which 115 are classified as plant cell wall degrading enzymes, which can act in fractions of hemicellulose and pectin. Our study highlights the potential of this Chitinophaga isolate provides several plant-polysaccharide-degrading enzymes.






Similar content being viewed by others
Data availability
The 16S rRNA gene sequences of isolates Chitinophaga sp. CB10 (CB10_01), P. nosoerga CB10 (CB10_02) and Labrys sp. CB10 (CB10_03) have been deposited in the GenBank database under accession numbers MW703979, MW703978 and MW703980, respectively. The genome sequences were deposited in GenBank database at the National Center for Biotechnology Information (NCBI) under the accession numbers: MLAV00000000 (Chitinophaga sp. CB10), NJGW00000000 (P. nosoerga CB10) and NJGX00000000 (Labrys sp. CB10). The raw data generated for this study were deposited in the NCBI - Sequence Read Archive (SRA), under BioProject ID PRJNA335650.
References
Alneberg J, Bjarnason BS, de Bruijn I et al (2014) Binning metagenomic contigs by coverage and composition. Nat Methods 11:1144–1146. https://doi.org/10.1038/nmeth.3103
Amorim CL, Moreira IS, Maia AS et al (2014) Biodegradation of ofloxacin, norfloxacin, and ciprofloxacin as single and mixed substrates by Labrys portucalensis F11. Appl Microbiol Biotechnol 98:3181–3190. https://doi.org/10.1007/s00253-013-5333-8
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge
Bowers RM, Kyrpides NC, Stepanauskas R et al (2017) Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. https://doi.org/10.1038/nbt.3893
Brethauer S, Studer MH (2015) Biochemical conversion processes of lignocellulosic biomass to fuels and chemicals—a review. Chimia 69:572–581. https://doi.org/10.2533/chimia.2015.572
Calusinska M, Marynowska M, Bertucci M et al (2020) Integrative omics analysis of the termite gut system adaptation to Miscanthus diet identifies lignocellulose degradation enzymes. Commun Biol 3:275. https://doi.org/10.1038/s42003-020-1004-3
Campos E, Negro Alvarez MJ, Sabarís di Lorenzo G et al (2014) Purification and characterization of a GH43 β-xylosidase from Enterobacter sp. identified and cloned from forest soil bacteria. Microbiol Res 169:213–220. https://doi.org/10.1016/j.micres.2013.06.004
Chen C-Y, Kuo J-T, Cheng C-Y et al (2009) Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system. J Hazard Mater 172:1439–1445. https://doi.org/10.1016/j.jhazmat.2009.08.009
Chettri D, Verma AK, Verma AK (2020) Innovations in CAZyme gene diversity and its modification for biorefinery applications. Biotechnol Rep 28:e00525. https://doi.org/10.1016/j.btre.2020.e00525
Chou Y-J, Elliott GN, James EK et al (2007) Labrys neptuniae sp. nov., isolated from root nodules of the aquatic legume Neptunia oleracea. Int J Syst Evol Microbiol 57:577–581. https://doi.org/10.1099/ijs.0.64553-0
Coenye T, Falsen E, Hoste B et al (2000) Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol 50:887–899. https://doi.org/10.1099/00207713-50-2-887
Constancio MTL, Sacco LP, Campanharo JC et al (2020) Exploring the potential of two bacterial consortia to degrade cellulosic biomass for biotechnological applications. Curr Microbiol 77:3114–3124. https://doi.org/10.1007/s00284-020-02136-7
Costessi A, van den Bogert B, May A et al (2018) Novel sequencing technologies to support industrial biotechnology. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny103
DeAngelis KM, Gladden JM, Allgaier M et al (2010) Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. Bioenergy Res 3:146–158. https://doi.org/10.1007/s12155-010-9089-z
Deloger M, El Karoui M, Petit M-A (2009) A genomic distance based on MUM indicates discontinuity between most bacterial species and genera. J Bacteriol 191:91–99. https://doi.org/10.1128/JB.01202-08
Didion JP, Martin M, Collins FS (2017) Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ 5:e3720. https://doi.org/10.7717/peerj.3720
Dong S, Chang Y, Shen J et al (2017) Purification, expression and characterization of a novel α- l -fucosidase from a marine bacteria Wenyingzhuangia fucanilytica. Protein Expr Purif 129:9–17. https://doi.org/10.1016/j.pep.2016.08.016
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Ewing B, Hillier L, Wendl MC et al (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185. https://doi.org/10.1101/gr.8.3.175
Fernandes GC, Sierra EGM, Brear P et al (2021) From data mining of Chitinophaga sp. genome to enzyme discovery of a hyperthermophilic metallocarboxypeptidase. Microorganisms 9:393. https://doi.org/10.3390/microorganisms9020393
Gao F, Zhang C-T (2008) Ori-finder: a web-based system for finding oriC s in unannotated bacterial genomes. BMC Bioinform 9:79. https://doi.org/10.1186/1471-2105-9-79
Gharechahi J, Salekdeh GH (2018) A metagenomic analysis of the camel rumen’s microbiome identifies the major microbes responsible for lignocellulose degradation and fermentation. Biotechnol Biofuels 11:216. https://doi.org/10.1186/s13068-018-1214-9
Gharechahi J, Vahidi MF, Bahram M et al (2020) Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. https://doi.org/10.1038/s41396-020-00837-2
Glavina Del Rio T, Abt B, Spring S et al (2010) Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034T). Stand Genomic Sci 2:87–95. https://doi.org/10.4056/sigs.661199
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202. https://doi.org/10.1101/gr.8.3.195
Goris J, Konstantinidis KT, Klappenbach JA et al (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Herlet J, Schwarz WH, Zverlov VV et al (2018) Addition of β-galactosidase boosts the xyloglucan degradation capability of endoglucanase Cel9D from Clostridium thermocellum. Biotechnol Biofuels 11:238. https://doi.org/10.1186/s13068-018-1242-5
Himmel ME, Xu Q, Luo Y et al (2010) Microbial enzyme systems for biomass conversion: emerging paradigms. Biofuels 1:323–341. https://doi.org/10.4155/bfs.09.25
Huang S, Sheng P, Zhang H (2012) Isolation and identification of cellulolytic bacteria from the gut of Holotrichia parallela larvae (Coleoptera: Scarabaeidae). IJMS 13:2563–2577. https://doi.org/10.3390/ijms13032563
Huelsenbeck JP, Ronquist F (2001) MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Huerta-Cepas J, Forslund K, Coelho LP et al (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Huerta-Cepas J, Szklarczyk D, Heller D et al (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. https://doi.org/10.1093/nar/gky1085
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. https://doi.org/10.1093/molbev/msj030
IMG/M Data Consortium, Nayfach S, Roux S et al (2020) A genomic catalog of earth’s microbiomes. Nat Biotechnol. https://doi.org/10.1038/s41587-020-0718-6
Janda JM, Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. https://doi.org/10.1128/JCM.01228-07
Jin Y, Zhou J, Zhou J et al (2020) Genome-based classification of Burkholderia cepacia complex provides new insight into its taxonomic status. Biol Direct 15:6. https://doi.org/10.1186/s13062-020-0258-5
Kandler O, Colwell RR, Krichevsky MI et al (1987) Report of the Ad Hoc Committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464. https://doi.org/10.1099/00207713-37-4-463
Kang DD, Li F, Kirton E et al (2019) MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. https://doi.org/10.7717/peerj.7359
Kishi LT, Lopes EM, Fernandes CC et al (2017) Draft genome sequence of a Chitinophaga strain isolated from a lignocellulose biomass-degrading consortium. Genome Announc 5:e01056-e1116. https://doi.org/10.1128/genomeA.01056-16
Kumar M, Verma S, Gazara RK et al (2018) Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating β-proteobacterium Pandoraea sp. ISTKB Biotechnol Biofuels 11:154. https://doi.org/10.1186/s13068-018-1148-2
Lander ES, Waterman MS (1988) Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231–239. https://doi.org/10.1016/0888-7543(88)90007-9
Langmead B, Trapnell C, Pop M et al (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. https://doi.org/10.1186/gb-2009-10-3-r25
Larsbrink J, Izumi A, Ibatullin FM et al (2011) Structural and enzymatic characterization of a glycoside hydrolase family 31 α-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem J 436:567–580. https://doi.org/10.1042/BJ20110299
Larsbrink J, Tuveng TR, Pope PB et al (2017) Proteomic insights into mannan degradation and protein secretion by the forest floor bacterium Chitinophaga pinensis. J Proteomics 156:63–74. https://doi.org/10.1016/j.jprot.2017.01.003
Lee CC, Kibblewhite RE, Wagschal K et al (2012) Isolation and characterization of a novel GH67 α-glucuronidase from a mixed culture. J Ind Microbiol Biotechnol 39:1245–1251. https://doi.org/10.1007/s10295-012-1128-7
Lee I, Ouk Kim Y, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. https://doi.org/10.1099/ijsem.0.000760
Leonel TF, Pepe ESG, Castellane TCL et al (2021) Bagasse minority pathway expression: real time study of GH2 β-mannosidases from bacteroidetes. PLoS ONE 16:e0247822. https://doi.org/10.1371/journal.pone.0247822
Li R, Zheng J-W, Ni B et al (2011) Biodegradation of pentachloronitrobenzene by Labrys portucalensis pcnb-21 isolated from polluted soil. Pedosphere 21:31–36. https://doi.org/10.1016/S1002-0160(10)60076-8
Li Q, Wu T, Qi Z et al (2018) Characterization of a novel thermostable and xylose-tolerant GH 39 β-xylosidase from Dictyoglomus thermophilum. BMC Biotechnol 18:29. https://doi.org/10.1186/s12896-018-0440-3
Liu N, Li H, Chevrette MG et al (2019) Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J 13:104–117. https://doi.org/10.1038/s41396-018-0255-1
Lombard V, Golaconda Ramulu H, Drula E et al (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490-495. https://doi.org/10.1093/nar/gkt1178
Luo H, Gao F (2019) DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucleic Acids Res 47:D74–D77. https://doi.org/10.1093/nar/gky1014
Malgas S, Mafa MS, Mkabayi L et al (2019) A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J Microbiol Biotechnol 35:187. https://doi.org/10.1007/s11274-019-2765-z
Marçais G, Kingsford C (2011) A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27:764–770. https://doi.org/10.1093/bioinformatics/btr011
McKee LS, Brumer H (2015) Growth of Chitinophaga pinensis on plant cell wall glycans and characterisation of a glycoside hydrolase family 27 \beta-l-arabinopyranosidase implicated in arabinogalactan utilisation. PLoS ONE 10:e0139932. https://doi.org/10.1371/journal.pone.0139932
McKee LS, Martínez-Abad A, Ruthes AC et al (2018) Focused metabolism of \beta-glucans by the soil bacteroidetes species Chitinophaga pinensis. Appl Environ Microbiol 85:e02231–e02218. https://doi.org/10.1128/AEM.02231-18
Meier-Kolthoff JP, Auch AF, Klenk H-P et al (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. https://doi.org/10.1186/1471-2105-14-60
Mhuantong W, Charoensawan V, Kanokratana P et al (2015) Comparative analysis of sugarcane bagasse metagenome reveals unique and conserved biomass-degrading enzymes among lignocellulolytic microbial communities. Biotechnol Biofuels 8:16. https://doi.org/10.1186/s13068-015-0200-8
Mikheenko A, Saveliev V, Gurevich A (2016) MetaQUAST: evaluation of metagenome assemblies. Bioinformatics 32:1088–1090. https://doi.org/10.1093/bioinformatics/btv697
Montella S, Ventorino V, Lombard V et al (2017) Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci Rep 7:42623. https://doi.org/10.1038/srep42623
Nurk S, Meleshko D, Korobeynikov A et al (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. https://doi.org/10.1101/gr.213959.116
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. https://github.com/nylander/MrModeltest2
Omori WP, Pinheiro DG, Kishi LT et al (2019) Draft genome of Thermomonospora sp. CIT 1 (Thermomonosporaceae) and in silico evidence of its functional role in filter cake biomass deconstruction. Genet Mol Biol 42:145–150. https://doi.org/10.1590/1678-4685-gmb-2017-0376
Parks DH, Imelfort M, Skennerton CT et al (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. https://doi.org/10.1101/gr.186072.114
Passari AK, Mishra VK, Leo VV et al (2016) Phytohormone production endowed with antagonistic potential and plant growth promoting abilities of culturable endophytic bacteria isolated from Clerodendrum colebrookianum Walp. Microbiol Res 193:57–73. https://doi.org/10.1016/j.micres.2016.09.006
Peeters C, De Canck E, Cnockaert M et al (2019) Comparative genomics of Pandoraea, a genus enriched in xenobiotic biodegradation and metabolism. Front Microbiol 10:2556. https://doi.org/10.3389/fmicb.2019.02556
Pham TTM, Tu Y, Sylvestre M (2012) Remarkable ability of Pandoraea pnomenusa B356 biphenyl dioxygenase to metabolize simple flavonoids. Appl Environ Microbiol 78:3560–3570. https://doi.org/10.1128/AEM.00225-12
Prior P, Ailloud F, Dalsing BL et al (2016) Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17:90. https://doi.org/10.1186/s12864-016-2413-z
Pruitt KD, Tatusova T, Maglott DR (2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35:D61–D65. https://doi.org/10.1093/nar/gkl842
Ramasamy D, Mishra AK, Lagier J-C et al (2014) A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol 64:384–391. https://doi.org/10.1099/ijs.0.057091-0
Rhee MS, Sawhney N, Kim YS et al (2017) GH115 α-glucuronidase and GH11 xylanase from Paenibacillus sp. JDR-2: potential roles in processing glucuronoxylans. Appl Microbiol Biotechnol 101:1465–1476. https://doi.org/10.1007/s00253-016-7899-4
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Rocha J, Botelho J, Ksiezarek M et al (2020) Lactobacillus mulieris sp. nov., a new species of Lactobacillus delbrueckii group. Int J Syst Evol Microbiol 70:1522–1527. https://doi.org/10.1099/ijsem.0.003901
Ryabova O, Vršanská M, Kaneko S et al (2009) A novel family of hemicellulolytic α-glucuronidase. FEBS Lett 583:1457–1462. https://doi.org/10.1016/j.febslet.2009.03.057
Rytioja J, Hildén K, Yuzon J et al (2014) Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol Mol Biol Rev 78:614–649. https://doi.org/10.1128/MMBR.00035-14
Sahin N, Tani A, Kotan R et al (2011) Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int J Syst Evol Microbiol 61:2247–2253. https://doi.org/10.1099/ijs.0.026138-0
Sangkhobol V, Skerman VBD (1981) Chitinophaga, a new genus of chitinolytic myxobacteria. Int J Syst Bacteriol 31:285–293. https://doi.org/10.1099/00207713-31-3-285
Sarhan MS, Hamza MA, Youssef HH et al (2019) Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media—a review. J Adv Res 19:15–27. https://doi.org/10.1016/j.jare.2019.04.002
Sawhney N, Preston JF (2014) GH51 arabinofuranosidase and its role in the methylglucuronoarabinoxylan utilization system in Paenibacillus sp. strain JDR-2. Appl Environ Microbiol 80:6114–6125. https://doi.org/10.1128/AEM.01684-14
Schmieder R, Edwards R (2011) Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. https://doi.org/10.1093/bioinformatics/btr026
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Seemann T (2018) barrnap 0.9 : rapid ribosomal RNA prediction. Program distributed by the author. https://github.com/tseemann/barrnap
Shi Y, Chai L, Tang C et al (2013) Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst Eng 36:1957–1965. https://doi.org/10.1007/s00449-013-0972-9
Shoseyov O, Shani Z, Levy I (2006) Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70:283–295. https://doi.org/10.1128/MMBR.00028-05
Sindhu R, Binod P, Pandey A (2016) Biological pretreatment of lignocellulosic biomass—an overview. Biores Technol 199:76–82. https://doi.org/10.1016/j.biortech.2015.08.030
Sweeney MD, Xu F (2012) Biomass converting enzymes as industrial biocatalysts for fuels and chemicals: recent developments. Catalysts 2:244–263. https://doi.org/10.3390/catal2020244
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Talamantes D, Biabini N, Dang H et al (2016) Natural diversity of cellulases, xylanases, and chitinases in bacteria. Biotechnol Biofuels 9:133. https://doi.org/10.1186/s13068-016-0538-6
Taylor-Brown A, Spang L, Borel N et al (2017) Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus chlamydia. Sci Rep 7:10661. https://doi.org/10.1038/s41598-017-10757-5
Uritskiy GV, DiRuggiero J, Taylor J (2018) MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. https://doi.org/10.1186/s40168-018-0541-1
Van Dyk JS, Pletschke BI (2012) A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—factors affecting enzymes, conversion and synergy. Biotechnol Adv 30:1458–1480. https://doi.org/10.1016/j.biotechadv.2012.03.002
Vurture GW, Sedlazeck FJ, Nattestad M et al (2017) GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33:2202–2204. https://doi.org/10.1093/bioinformatics/btx153
Wang Q, Garrity GM, Tiedje JM et al (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Weisburg WG, Barns SM, Pelletier DA et al (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. https://doi.org/10.1128/JB.173.2.697-703.1991
Wu Y-W, Tang Y-H, Tringe SG et al (2014) MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26. https://doi.org/10.1186/2049-2618-2-26
Yu J, Cai W, Cheng Z, Chen J (2014) Degradation of dichloromethane by an isolated strain Pandoraea pnomenusa and its performance in a biotrickling filter. J Environ Sci 26:1108–1117. https://doi.org/10.1016/S1001-0742(13)60538-0
Zhang H, Yohe T, Huang L et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. https://doi.org/10.1093/nar/gky418
Acknowledgements
The authors would like to acknowledge the Programa de Pós-Graduação em Microbiologia Agropecuária, Universidade Estadual Paulista (UNESP), Faculdade de Ciências Agrárias e Veterinárias, Jaboticabal, SP, Brazil, for the academic support. The authors thank to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the scholarships of M.I.G.F. and L.A.L.C.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 and Conselho Nacional de desenvolvimento Científico e Tecnológico (CNPq). This work also was supported by grants from the São Paulo Research Foundation (FAPESP) [Nos. 2010/17520-9 and 2016/16624-1].
Author information
Authors and Affiliations
Contributions
Conceptualization: MIGF, DGP, LTK, EGML; methodology: MIGF, DGP; formal analysis: MIGF, DGP, ESGP, LALC, JCC, CCF; investigation: MIGF, DGP; writing - original draft preparation: MIGF; writing - review and editing: DGP, ESGP, LALC; JCC, CCF, LTK, LMCA, EGML; funding acquisition: EGML, LMCA; LTK; resources: EGML, LMCA; supervision: DGP, EGML.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11274_2021_3128_MOESM6_ESM.eps
Supplementary file6 Qualitative cellulase activity assay. Cultures grown on a plate containing Avicel (PH-101) before (a,b,c,d), and after (e,f,g,h) staining with Congo red, (a,e) Chitinophaga sp. (isolate CB10_01) (b,f) Pandoraea nosoerga (isolate CB10_02) (c,g), Labrys sp. (isolate CB10_03), and (d,h) CB10 community (EPS 63490 kb)
11274_2021_3128_MOESM7_ESM.eps
Supplementary file7 Genomic location of the predicted oriC site (highlighted in black) in Chitinophaga sp. CB10 compared to the Chitinophaga pinensis DSM 2588 genome available in DoriC database. The similarities between genomes are represented by the gray bands (EPS 84 kb)
11274_2021_3128_MOESM8_ESM.eps
Supplementary file8 Genomic location of the predicted oriC site (highlighted in black) in Pandoraea nosoerga CB10 compared to the Pandoraea pulmonicola DSM 16583 genome available in DoriC database. The similarities between genomes are represented by the gray bands (EPS 70 kb)
11274_2021_3128_MOESM9_ESM.eps
Supplementary file9 Consensus network tree derived from Neighbor-Joining trees from genomic distances of the Chitinophaga genus (EPS 10406 kb)
11274_2021_3128_MOESM10_ESM.eps
Supplementary file10 Consensus network tree derived from Neighbor-Joining trees from genomic distances of the Pandoraea genus (EPS 10724 kb)
Rights and permissions
About this article
Cite this article
Funnicelli, M.I.G., Pinheiro, D.G., Gomes-Pepe, E.S. et al. Metagenome-assembled genome of a Chitinophaga sp. and its potential in plant biomass degradation, as well of affiliated Pandoraea and Labrys species. World J Microbiol Biotechnol 37, 162 (2021). https://doi.org/10.1007/s11274-021-03128-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03128-w