Abstract
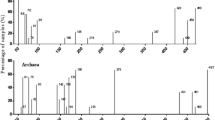
Study of rhizospheric bacteria from important plants is very essential, as they are known to influence plant growth and productivity, and also produce industrially important metabolites. Origanum vulgare is a perennial medicinal aromatic plant rich in phenolic antioxidants. Present study investigates the diversity of culturable root-associated bacteria from this plant in Palampur, India, which constitutes a unique ecosystem due to high rain fall, wide temperature fluctuations and acidic soil. Both root endophytes and rhizospheric soil bacteria were isolated, which resulted in a total of 120 morphologically different isolates. They were found to group into 21 phylotypes based on restriction fragment length polymorphism analysis. Growth medium composition had significant effect on the diversity of the isolated bacterial populations. The isolates were characterized for various metabolic, plant growth promoting (PGP) and other biotechnologically useful activities, based on which they were clustered into groups by principal component analysis. Majority of the isolates belonged to γ-Proteobacteria and Firmicutes. Pseudomonas and Stenotrophomonas were the most dominant species and together constituted 27.5 % of the total isolates. Many isolates, especially γ-Proteobacteria, showed very high PGP activities. Few isolates exhibited very high antioxidant activity, which may find potential applications in food and health industries. Firmicutes were catabolically the most versatile group and produced several hydrolytic enzymes. To the best of our knowledge, it is the first study describing rhizospheric microbial community of O. vulgare.







Similar content being viewed by others
References
Afifi AF (1978) Effect of volatile substances released from Origanum majorana and Ocimum basilicum on the rhizosphere and phyllosphere fungi of Phaseolus vulgaris. Folia Microbiol 23:399–405
Babalola OO (2010) Beneficial bacteria of agricultural importance. Biotechnol Lett 32:1559–1570
Banchio E, Bogino PC, Zygadlo J, Giordano W (2008) Plant growth promoting rhizobacteria improve growth and essential oil yield in Origanum majorana L. Biochem Syst Ecol 36:766–771
Bednarek P, Kwon C, Schulze-Lefert P (2010) Not a peripheral issue: secretion in plant-microbe interactions. Curr Opin Plant Biol 13:378–387
Bergey DH, Holt JG (1994) Bergey’s manual of determinative bacteriology, 9th edn. Lippincott Williams and Wilkins, Baltimore
Çakmakçı R, Dönmez MF, Ertürk Y, Erat M, Haznedar A, Sekban R (2010) Diversity and metabolic potential of culturable bacteria from the rhizosphere of Turkish tea grown in acidic soils. Plant Soil 332:299–318
Chauhan PS, Chaudhry V, Mishra S, Nautiyal CS (2011) Uncultured bacterial diversity in tropical maize (Zea mays L.) rhizosphere. J Basic Microbiol 51:15–32
Chou TH, Ding HY, Hung WJ, Liang CH (2010) Antioxidative characteristics and inhibition of α-melanocyte-stimulating hormone-stimulated melanogenesis of vanillin and vanillic acid from Origanum vulgare. Exp Dermatol 19:742–750
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(suppl 1):D141–D145
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package), version 3.6a3. Distributed by the author. Department of Genetics, University of Washington, Seattle
Gardener BBM, Weller DM (2001) Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl Environ Microbiol 67:4414–4425
Gerhardt P, Murray RGE, Costilov RN, Nester EW, Wood WA, Kreig NR, Philips GB (1981) Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC
Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3:408–414
Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230
Haldar S, Choudhury SR, Sengupta S (2011) Genetic and functional diversities of bacterial communities in the rhizosphere of Arachis hypogaea. Antonie Van Leeuwenhoek 100:161–170
Hayes LA, Lovley DR (2002) Specific 16S rDNA sequences associated with naphthalene degradation under sulfate-reducing conditions in harbor sediments. Microb Ecol 43:134–145
Kumar G, Kanaujia N, Bafana A (2012) Functional and phylogenetic diversity of root-associated bacteria of Ajuga bracteosa in Kangra valley. Microbiol Res 167:220–225
Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, Piceno YM, DeSantis TZ, Andersen GL, Bakker PAHM, Raaijmakers JM (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Mishra PK, Bisht SC, Ruwari P, Selvakumar G, Joshi GK, Bisht JK, Bhatt JC, Gupta HS (2011) Alleviation of cold stress in inoculated wheat (Triticum aestivum L.) seedlings with psychrotolerant Pseudomonads from NW Himalayas. Arch Microbiol 193:497–513
Oyaizu M (1986) Studies on products of browning reaction prepared from glucosamine. Jpn J Nutr 44:307–314
Page AL, Miller RH, Keeney DR (1982) Method of soil analysis. Part 2, chemical and microbiological properties of soils, 2nd edn. American Society of Agronomy, Soil Science Society of America, Madison
Pande C, Tewari G, Singh S, Singh C (2012) Chemical markers in Origanum vulgare L. from Kumaon Himalayas: a chemosystematic study. Nat Prod Res 26:140–145
Pandey A, Palni LMS (1996) The rhizosphere effect of tea on soil microbes in a Himalayan monsoonal location. Biol Fertil Soils 21:131–137
Pongsilp N, Nimnoi P, Lumyong S (2012) Genotypic diversity among rhizospheric bacteria of three legumes assessed by cultivation-dependent and cultivation-independent techniques. World J Microbiol Biotechnol 28:615–626
Qi X, Wang E, Xing M, Zhao W, Chen X (2012) Rhizosphere and non-rhizosphere bacterial community composition of the wild medicinal plant Rumex patientia. World J Microbiol Biotechnol 28:2257–2265
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, USA
Senechkin IV, Speksnijder AGCL, Semenov AM, van Bruggen AHC, van Overbeekm LS (2010) Isolation and partial characterization of bacterial strains on low organic carbon medium from soils fertilized with different organic amendments. Microb Ecol 60:829–839
Sood A, Sharma S, Kumar V, Thakur RL (2008) Established and abandoned tea (Camillia sinensis L.) rhizosphere: dominant bacteria and their antagonism. Pol J Microbiol 57:71–76
Strycharz S, Shetty K (2002) Response of oregano (Origanum vulgare L.) clonal lines to Pseudomonas sp. Z strain and polydye R-478 and implications for hyperhydricity prevention in tissue culture. Process Biochem 38:343–350
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb Ecol 62:324–336
Tsavkelova EA, Cherdyntseva TA, Klimova SY, Shestakov AI, Botina SG, Netrusov AI (2007) Orchid-associated bacteria produce indole-3-acetic acid, promote seed germination, and increase their microbial yield in response to exogenous auxin. Arch Microbiol 188:655–664
Verma RS, Padalia RC, Chauhan A, Verma RK, Yadav AK, Singh HP (2010) Chemical diversity in Indian oregano (Origanum vulgare L.). Chem Biodivers 7:2054–2064
Acknowledgments
The author is thankful to Director, NERRI, Nagpur, and Director, IHBT, Palampur, for providing necessary facilities to carry out the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bafana, A. Diversity and metabolic potential of culturable root-associated bacteria from Origanum vulgare in sub-Himalayan region. World J Microbiol Biotechnol 29, 63–74 (2013). https://doi.org/10.1007/s11274-012-1158-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-012-1158-3