Abstract
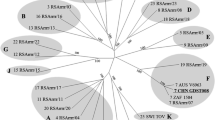
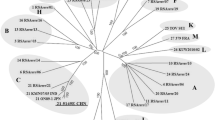
Bluetongue disease is a reportable animal disease that affects wild and farmed ruminants, including white-tailed deer (WTD). This report documents the clinical findings, ancillary diagnostics, and genomic characterization of a novel reassortant bluetongue virus serotype 2 (BTV-2) strain isolated from a dead Florida farmed WTD in 2022. Our analyses support that this BTV-2 strain likely stemmed from the acquisition of genome segments from co-circulating BTV strains in Florida and Louisiana. In addition, our analyses also indicate that genetically uncharacterized BTV strains may be circulating in the Southeastern USA; however, the identity and reassortant status of these BTV strains cannot be determined based on the VP2 and VP5 genome sequences. Hence, continued surveillance based on complete genome characterization is needed to understand the genetic diversity of BTV strains in this region and the potential threat they may pose to the health of deer and other ruminants.

Similar content being viewed by others
Data availability
The complete gene coding sequences for all 10 segments of the genomes recovered in this study have been deposited in the NCBI GenBank database and are available under the GenBank accession nos. OR672561-OR672570.
References
Erasmus BJ (1990) Bluetongue Virus. In: Dinter Z, Morein B (eds) virus infections of vertebrates series, 1st edn. Elsevier, Amsterdam, pp 227–237
Du Toit RM (1944) The Transmission of Blue-tongue and Horse-sickness by Culicoides. Onderstepoort J Vet Res 19(1–2):7–16
Mertens PPC, Diprose J (2004) The bluetongue virus core: a nano-scale transcription machine. Virus Res 101(1):29–43. https://doi.org/10.1016/j.virusres.2003.12.004
Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M (2011) Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog 7(12):e1002477. https://doi.org/10.1371/journal.ppat.1002477
Roy P, Urakawa T, Van Dijk AA, Erasmus BJ (1990) Recombinant virus vaccine for bluetongue disease in sheep. J Virol 64(5):1998–2003. https://doi.org/10.1128/JVI.64.5.1998-2003.1990
Hofmann MA, Renzullo S, Mader M, Chaignat V, Worwa G, Thuer B (2008) Genetic characterization of toggenburg orbivirus, a new bluetongue virus, from goats, Switzerland. Emerg Infect Dis 14:1855–1861. https://doi.org/10.3201/eid1412.080818
Lorusso A, Sghaier S, Di Domenico M, Barbria ME, Zaccaria G, Megdich A, Portanti O, Seliman IB, Spedicato M, Pizzurro F et al (2018) Analysis of bluetongue serotype 3 spread in Tunisia and discovery of a novel strain related to the bluetongue virus isolated from a commercial sheep pox vaccine. Infect Genet Evol 59:63–71. https://doi.org/10.1016/j.meegid.2018.01.025
Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek-Bankowska K, Belaganahalli MN, Attoui H, Mertens PPC (2011) Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS ONE 6:e26147. https://doi.org/10.1371/journal.pone.0026147
Marcacci M, Sant S, Mangone I, Goria M, Dondo A, Zoppi S, van Gennip RGP, Radaelli MC, Cammà C, van Rijn PA et al (2018) One after the other: A novel Bluetongue virus strain related to Toggenburg virus detected in the Piedmont region (North-western Italy), extends the panel of novel atypical BTV strains. Transbound Emerg Dis 65:370–374. https://doi.org/10.1111/tbed.12822
Savini G, Puggioni G, Meloni G, Marcacci M, Di Domenico M, Rocchigiani AM, Spedicato M, Oggiano A, Manunta D, Teodori L et al (2017) Novel putative Bluetongue virus in healthy goats from Sardinia, Italy. Infect Genet Evol 51:108–117. https://doi.org/10.1016/j.meegid.2017.03.021
Yang H, Gu W, Li Z, Zhang L, Liao D, Song J, Shi B, Hasimu J, Li Z, Yang Z et al (2021) Novel putative bluetongue virus serotype 29 isolated from inapparently infected goat in Xinjiang of China. Transbound Emerg Dis 68:2543–2555. https://doi.org/10.1111/tbed.13927
Zientara S, Sailleau C, Viarouge C, Höper D, Beer M, Jenckel M, Hoffmann B, Romey A, Bakkali-Kassimi L, Fablet A et al (2014) Novel bluetongue virus in goats, Corsica, France, 2014. Emerg Infect Dis 20:2123–2125. https://doi.org/10.3201/eid2012.140924
Bréard E, Sailleau C, Coupier H, Mure-Ravaud K, Hammoumi S, Gicquel B, Hamblin C, Dubourget P, Zientara S (2003) Comparison of genome segments 2, 7, and 10 of bluetongue viruses serotype 2 for differentiation between field isolates and the vaccine strain. Vet Res 34(6):777–789. https://doi.org/10.1051/vetres:2003036
Gaudreault NN, Mayo CE, Jasperson DC, Crossley BM, Breitmeyer RE, Johnson DJ, Ostlund EN, MacLachlan NJ, Wilson WC (2014) Whole genome sequencing and phylogenetic analysis of Bluetongue virus serotype 2 strains isolated in the Americas including a novel strain from the western United States. J Vet Diagn Invest 26(4):553–557. https://doi.org/10.1177/1040638714536902
Gibbs EP, Greiner EC, Taylor WP, Barber TL, House JA, Pearson JE (1983) Isolation of bluetongue virus serotype 2 from cattle in Florida: serotype of bluetongue virus hitherto unrecognized in the Western Hemisphere. Am J Vet Res 44(12):2226–2228
Kundlacz C, Caignard G, Sailleau C, Viarouge C, Postic L, Vitour D, Zientara S, Breard E (2019) Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 11(7):672. https://doi.org/10.3390/v11070672
Nomikou K, Hughes J, Wash R, Kellam P, Breard E, Zientara S, Palmarini M, Biek R, Mertens P (2015) Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion. PLOS Pathog 11(8):e1005056. https://doi.org/10.1371/journal.ppat.1005056
Greiner EC, Barber TL, Pearson JE, Kramer W, Gibbs EP (1985) Orbiviruses from culicoides in Florida. In: Barber TL, Jochum MM (eds) Bluetongue and related orbiviruses, 1st edn. Alan R. Liss, New York, pp 195–200
Wright JC, Lauerman LH, Nusbaum KE, Mullen GR (1989) Seroepidemiologic study of bluetongue virus serotype 2 in Alabama. Prev Vet Med 7(2):113–119. https://doi.org/10.1016/0167-5877(89)90003-2
Maclachlan NJ, Wilson WC, Crossley BM, Mayo CE, Jasperson DC, Breitmeyer RE, Whiteford AM (2013) Novel serotype of bluetongue virus, western North America. Emerg Infect Dis 19(4):665–666. https://doi.org/10.3201/eid1904.120347
USAHA (2014) Proceedings one hundred and eighteenth annual meeting of the United States animal health association. Kansas City, Missouri.
USAHA (2016). Proceedings one hundred and twentieth annual meeting of the United States animal health association. Greensboro, North Carolina.
USAHA (2020). Proceedings one hundred and twenty fourth annual meeting of the United States animal health association. Nashville, Tennessee (virtual meeting).
Ahasan MS, Subramaniam K, Sayler KA, Loeb JC, Popov VL, Lednicky JA, Wisely SM, Campos Krauer JM, Waltzek TB (2019) Molecular characterization of a novel reassortment Mammalian orthoreovirus type 2 isolated from a Florida white-tailed deer fawn. Virus Res 270:97642. https://doi.org/10.1016/j.virusres.2019.197642
Lambert AJ, Martin DA, Lanciotti RS (2003) Detection of North American eastern and western equine encephalitis viruses by nucleic acid amplification assays. J Clin Microbiol 41(1):379–385. https://doi.org/10.1128/JCM.41.1.379-385.2003
Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT (2000) Rapid detection of west nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38(11):4066–4071. https://doi.org/10.1128/JCM.38.11.4066-4071.2000
Wernike K, Hoffmann B, Beer M (2015) Simultaneous detection of five notifiable viral diseases of cattle by single-tube multiplex real-time RT-PCR. J Virol Methods 217:28–35. https://doi.org/10.1016/j.jviromet.2015.02.023
Sayler KA, Subramaniam K, Jacob JM, Loeb JC, Craft WF, Farina LL, Stacy NI, Moussatche N, Cook L, Lednicky JA, Wisely SM, Waltzek TB (2019) Characterization of mule deerpox virus in Florida white-tailed deer fawns expands the known host and geographic range of this emerging pathogen. Arch Virol 164(1):51–61. https://doi.org/10.1007/s00705-018-3991-7
Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15(3):R46. https://doi.org/10.1186/gb-2014-15-3-r46
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274. https://doi.org/10.1093/molbev/msu300
Caporale M, Di Gialleonorado L, Janowicz A, Wilkie G, Shaw A, Savini G, Van Rijn PA, Mertens P, Di Ventura M, Palmarini M (2014) Virus and host factors affecting the clinical outcome of bluetongue virus infection. J Virol 88:10399–10411. https://doi.org/10.1128/JVI.01641-14
Parsonson IM (1990) Pathology and pathogenesis of bluetongue infections. Curr Top Microbiol Immunol 162:119–141. https://doi.org/10.1007/978-3-642-75247-6_5
Rivera NA, Varga C, Ruder MG, Dorak SJ, Roca AL, Novakofski JE, Mateus-Pinilla NE (2021) Bluetongue and epizootic hemorrhagic disease in the United States of America at the wildlife-livestock interface. Pathog 10(8):915. https://doi.org/10.3390/pathogens10080915
Sellers RF, Maarouf AR (1989) Trajectory analysis and bluetongue virus serotype 2 in Florida 1982. Can J Vet Res 53(1):100–102
Alkhamis MA, Aguilar-Vega C, Fountain-Jones NM, Lin K, Perez AM, Sánchez-Vizcaíno JM (2020) Global emergence and evolutionary dynamics of bluetongue virus. Sci Rep 10:21677. https://doi.org/10.1038/s41598-020-78673-9
Funding
This research was funded by the University of Florida, Institute for Food and Agricultural Sciences, Cervidae Health Research Initiative, funded through the Florida State Legislature, grant number LBR2199.
Author information
Authors and Affiliations
Contributions
Conceptualization: PHOV, JAL, SMW, KS. Methodology: PHOV, JMCK, JAL, SMW, KS. Formal analysis: PHOV, AS, ACC, CED, SM, KNW, ZSW, ED, TDL, JMCK. Investigation: PHOV, AS, ACC, CED, SM, KNW, ZSW, ED, TDL, KS. Resources: JMCK, JAL, SMW, KS. Writing—original draft: PHOV, AS, JAL, KS. Writing—review & editing: PHOV, AS, ACC, CED, SM, KNW, ZSW, ED, TDL, JMCK, JAL, SMW, KS. Visualization: PHOV, AS, KS. Supervision: KS. Project administration: SMW, KS. Funding acquisition: SMW, KS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This work was approved by the University of Florida Institutional Animal Care and Use Committee (IACUC) Protocol Numbers 201609390 and 201909390.
Additional information
Edited by Juergen Richt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Viadanna, P.H.O., Surphlis, A., Cheng, AC. et al. A novel bluetongue virus serotype 2 strain isolated from a farmed Florida white-tailed deer (Odocoileus virginianus) arose from reassortment of gene segments derived from co-circulating serotypes in the Southeastern USA. Virus Genes 60, 100–104 (2024). https://doi.org/10.1007/s11262-023-02047-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-023-02047-2