Abstract
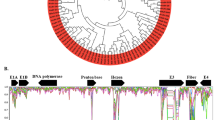
Human mastadenoviruses (HAdVs) are non-enveloped, double-stranded DNA viruses that are comprised of more than 85 types classified within seven species (A–G) based on genomics. All HAdV prototypes and many newly defined type genomes have been completely sequenced and are available. Computational analyses of the prototypes and newly emergent HAdV strains provide insights into the evolutionary history and molecular adaptation of HAdV. Most types of HAdV-B are important pathogens causing severe respiratory infections or urinary tract infections and are well characterized. However, HAdV-16 of the B1 subspecies has rarely been reported and its genome is poorly characterized. In this study, bioinformatics analysis, based on genome sequences obtained in GenBank, suggested that HAdV-16, a prototype HAdV-B species, evolved from multiple intertypic recombination events. HAdV-16 genome contains the hexon loop 1 to loop 2 region from HAdV-E4, the partial hexon conserved region 4 (C4) from the subspecies HAdV-B2, genome region 30,897–33,384 containing the fiber gene from SAdV-35, and other genomic parts from the subspecies HAdV-B1. Moreover, analysis of sequence similarity with HAdV-E4 LI, LII, and SAdV-36 strains demonstrated the recombination events happened rather early. Further, amino acid sequence alignment indicated that the amino acid variations occurred in hypervariable regions (HVRs). Especially, the major difference in HVR7, which contains the critical neutralization epitope of HAdV-E4, between HAdV-16 and HAdV-E4 might explain the low level of cross-neutralization between these strains. Our findings promote better understanding on HAdV evolution, predicting newly emergent HAdV strains, and developing novel HAdV vectors.



Similar content being viewed by others
References
Espinola EE, Barrios JC, Russomando G, Mirazo S, Arbiza J (2017) Computational analysis of a species D human adenovirus provides evidence of a novel virus. J Gen Virol 98(11):2810–2820
Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research C (2011) Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85:5701–5702
Yoshitomi H, Sera N, Gonzalez G, Hanaoka N, Fujimoto T (2017) First isolation of a new type of human adenovirus (genotype 79), species Human mastadenovirus B (B2) from sewage water in Japan. J Med Virol 89:1192–1200
Hashimoto S, Gonzalez G, Harada S, Oosako H, Hanaoka N, Hinokuma R, Fujimoto T (2018) Recombinant type Human mastadenovirus D85 associated with epidemic keratoconjunctivitis since 2015 in Japan. J Med Virol 90:881–889
Lenaerts L, De Clercq E, Naesens L (2008) Clinical features and treatment of adenovirus infections. Rev Med Virol 18:357–374
Sandkovsky U, Vargas L, Florescu DF (2014) Adenovirus: current epidemiology and emerging approaches to prevention and treatment. Curr Infect Dis Rep 16:416
Walsh MP, Seto J, Jones MS, Chodosh J, Xu W, Seto D (2010) Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J Clin Microbiol 48:991–993
Yang Z, Zhu Z, Tang L, Wang L, Tan X, Yu P, Zhang Y, Tian X, Wang J, Zhang Y, Li D, Xu W (2009) Genomic analyses of recombinant adenovirus type 11a in China. J Clin Microbiol 47:3082–3090
Dehghan S, Seto J, Liu EB, Walsh MP, Dyer DW, Chodosh J, Seto D (2013) Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443:197–207
Iampietro MJ, Larocca RA, Provine NM, Abbink P, Kang ZH, Bricault CA, Barouch DH (2018) Immunogenicity and cross-reactivity of rhesus adenoviral vectors. J Virol 92:e00159-18
Michael NL (2012) Rare serotype adenoviral vectors for HIV vaccine development. J Clin Invest 122:25–27
Madisch I, Harste G, Pommer H, Heim A (2005) Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J Virol 79:15265–15276
Pring-Akerblom P, Trijssenaar FE, Adrian T (1995) Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology 212:232–236
Ebner K, Pinsker W, Lion T (2005) Comparative sequence analysis of the hexon gene in the entire spectrum of human adenovirus serotypes: phylogenetic, taxonomic, and clinical implications. J Virol 79:12635–12642
Rafajko RR (1964) Production and standardization of adenovirus types 1 to 18 reference antisera. Am J Hyg 79:310–319
Wigand R (1987) Pitfalls in the identification of adenoviruses. J Virol Methods 16:161–169
Hierholzer JC, Stone YO, Broderson JR (1991) Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch Virol 121:179–197
Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, Blaney KM, Ligler AG, Malanoski AP, Santiago J, Walter EA, Agan BK, Metzgar D, Seto D, Daum LT, Kruzelock R, Rowley RK, Hanson EH, Tibbetts C, Stenger DA (2006) Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res 16:527–535
Tcherepanov V, Ehlers A, Upton C (2006) Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150
Lefort V, Longueville JE, Gascuel O (2017) SMS: smart Model Selection in PhyML. Mol Biol Evol 34:2422–2424
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225
Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Martin DP, Posada D, Crandall KA, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrovir 21:98–102
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E (1996) Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol 70:2116–2123
Roy S, Clawson DS, Calcedo R, Lebherz C, Sanmiguel J, Wu D, Wilson JM (2005) Use of chimeric adenoviral vectors to assess capsid neutralization determinants. Virology 333:207–214
Sumida SM, Truitt DM, Lemckert AA, Vogels R, Custers JH, Addo MM, Lockman S, Peter T, Peyerl FW, Kishko MG, Jackson SS, Gorgone DA, Lifton MA, Essex M, Walker BD, Goudsmit J, Havenga MJ, Barouch DH (2005) Neutralizing antibodies to adenovirus serotype 5 vaccine vectors are directed primarily against the adenovirus hexon protein. J Immunol 174:7179–7185
Tian X, Qiu H, Zhou Z, Wang S, Fan Y, Li X, Chu R, Li H, Zhou R, Wang H (2018) Identification of a critical and conformational neutralizing epitope in human adenovirus type 4 hexon. J Virol 92:e01643-17
Wu H, Dmitriev I, Kashentseva E, Seki T, Wang M, Curiel DT (2002) Construction and characterization of adenovirus serotype 5 packaged by serotype 3 hexon. J Virol 76:12775–12782
Yu B, Dong J, Wang C, Zhan Y, Zhang H, Wu J, Kong W, Yu X (2013) Characteristics of neutralizing antibodies to adenovirus capsid proteins in human and animal sera. Virology 437:118–123
Tian X, Su X, Li H, Li X, Zhou Z, Liu W, Zhou R (2011) Construction and characterization of human adenovirus serotype 3 packaged by serotype 7 hexon. Virus Res 160:214–220
Bradley RR, Maxfield LF, Lynch DM, Iampietro MJ, Borducchi EN, Barouch DH (2012) Adenovirus serotype 5-specific neutralizing antibodies target multiple hexon hypervariable regions. J Virol 86:1267–1272
Pichla-Gollon SL, Drinker M, Zhou X, Xue F, Rux JJ, Gao GP, Wilson JM, Ertl HC, Burnett RM, Bergelson JM (2007) Structure-based identification of a major neutralizing site in an adenovirus hexon. J Virol 81:1680–1689
Qiu H, Li X, Tian X, Zhou Z, Xing K, Li H, Tang N, Liu W, Bai P, Zhou R (2012) Serotype-specific neutralizing antibody epitopes of human adenovirus type 3 (HAdV-3) and HAdV-7 reside in multiple hexon hypervariable regions. J Virol 86:7964–7975
Rux JJ, Kuser PR, Burnett RM (2003) Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution x-ray crystallographic, molecular modeling, and sequence-based methods. J Virol 77:9553–9566
Tian X, Ma Q, Jiang Z, Huang J, Liu Q, Lu X, Luo Q, Zhou R (2015) Identification and application of neutralizing epitopes of human adenovirus type 55 hexon protein. Viruses 7:5632–5642
Yuan X, Qu Z, Wu X, Wang Y, Liu L, Wei F, Gao H, Shang L, Zhang H, Cui H, Zhao Y, Wu N, Tang Y, Qin L (2009) Molecular modeling and epitopes mapping of human adenovirus type 3 hexon protein. Vaccine 27:5103–5110
Li QG, Wadell G (1988) The degree of genetic variability among adenovirus type 4 strains isolated from man and chimpanzee. Arch Virol 101:65–77
Kajon AE, Lamson DM, Bair CR, Lu X, Landry ML, Menegus M, Erdman DD, St. George K (2018) Adenovirus type 4 respiratory infections among civilian adults, Northeastern United States, 2011-2015(1). Emerg Infect Dis 24:201–209
Kajon AE, Dickson LM, Murtagh P, Viale D, Carballal G, Echavarria M (2010) Molecular characterization of an adenovirus 3-16 intertypic recombinant isolated in Argentina from an infant hospitalized with acute respiratory infection. J Clin Microbiol 48:1494–1496
Youil R, Toner TJ, Su Q, Chen M, Tang A, Bett AJ, Casimiro D (2002) Hexon gene switch strategy for the generation of chimeric recombinant adenovirus. Hum Gene Ther 13:311–320
Acknowledgements
This study was supported by grants from the National Key Research and Development Program of China (2018YFC1200100), National Natural Science Foundation of China (NSFC 31570163), and the Youth Project of State Key Laboratory of Respiratory Disease (SKLRD-QN-201713). We sincerely thank the reviewers for their comments, which have greatly improved the quality of this study.
Author information
Authors and Affiliations
Contributions
XT conceived and wrote the paper, HW performed most computational analyses and revised the paper, and RZ provided advice and supervised the work. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Edited by Detlev H. Kruger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tian, X., Wu, H. & Zhou, R. Molecular evolution of human adenovirus type 16 through multiple recombination events. Virus Genes 55, 769–778 (2019). https://doi.org/10.1007/s11262-019-01698-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-019-01698-4