Abstract
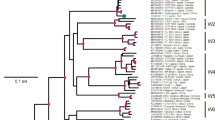
Apple mosaic virus (ApMV) is a widespread ssRNA virus which infects diverse species of Rosales. The phylogenetic analysis of complete capsid protein gene of the largest set of ApMV isolates discriminated two main clusters of isolates: one cluster correlates with Maloideae hosts and Trebouxia lichen algae hosts; a second with hop, Prunus, and other woody tree hosts. No correlation was found between clusters and geographic origin of virus isolates, and positive selection hypothesis in distinct hosts was not confirmed: in all virus populations, purifying selection had occurred. GGT→AAT substitution resulted in Gly→Asn change inside the zinc-finger motif in the capsid protein was revealed specific for discrimination of the clusters and we hypothesise that could influence the host preference.

Similar content being viewed by others
References
R.P. White, Plant Dis. Rep. 12, 33–34 (1928)
J. Bujarski, M. Figlerowicz, D. Gallitelli, M.J. Roossinck, S.W. Scott, Genus Ilarvirus. in Virus taxonomy, Classification and nomenclature of viruses, eds by A.M.Q. King, M.J. Adams, E.B. Carstens, E.J. Lefkowitz. Ninth Report of the International Committee on Taxonomy of Viruses (Elsevier Academic Press, Amsterdam, 2012) pp. 972–975
CAB International, Crop Protection Compendium (CAB International, Wallingford, 2003)
I.E. Tzanetakis, R. Martin, Plant Dis. 89, 431 (2005)
K. Petrzik, J. Vondrák, M. Barták, O. Peksa, O. Kubešová, Eur. J. Plant Pathol. (2013, in press)
S. Paunovic, G. Pasquini, M. Barba, Apple mosaic virus in stone fruits, in Virus and virus-like diseases of pome and stone fruits, ed. by A. Hadidi, M. Barba, T. Candresse, W. Jelkmann (APS Press, St. Paul, 2011), pp. 91–95
D.R. Crowle, S.J. Pethybridge, G.W. Leggett, L.J. Sherriff, C.R. Wilson, Plant Pathol. 52, 655–662 (2003)
R.W. Fulton, Apple mosaic virus. CMI/AAB Descriptions of plant viruses, 83 (Association of Applied Biologists, Wellesbourne, 1972)
A. Hadidi, M. Barba, Economic impact of pome and stone fruit viruses and viroids, in Virus and virus-like diseases of pome and stone fruits, ed. by A. Hadidi, M. Barba, T. Candresse, W. Jelkmann (APS Press, St. Paul, 2011), pp. 1–7
A.J. Gibbs, D. Fargette, F. García-Arenal, M.J. Gibbs, J. Gen. Virol. 91, 13–22 (2010)
K. Petrzik, Eur. J. Plant Pathol. 111, 355–360 (2005)
K. Petrzik, O. Lenz, Arch. Virol. 147, 1275–1285 (2002)
V. Lakshmi, V. Hallan, R. Ram, N. Ahmed, A.A. Zaidi, A. Varma, Indian J. Virol. 22, 44–49 (2011)
C. Desbiez, B. Moury, H. Lecoq, Infect. Genet. Evol. 11, 812–824 (2011)
R. Sanjuán, PLoS Pathog. 8(5), e1002685 (2012)
S.F. Elena, P. Agudelo-Romero, P. Carrasco, F.N. Codoner, S. Martín, C. Torres-Barceló, R. Sanjuán, Heredity 100, 478–483 (2008)
E.A.G. van der Vossen, L. Neeleman, J.F. Bol, Virology 202, 891–903 (1994)
P. Ansel-McKinney, S.W. Scott, M. Swanson, X. Ge, L. Gehrke, EMBO J. 15, 5077–5084 (1996)
P.C. Sehnke, A.M. Mason, S.J. Hood, R.M. Lister, J.E. Johnson, Virology 168, 48–56 (1989)
J.A. Sánchez-Navarro, V. Pallás, J. Gen. Virol. 75, 1441–1445 (1994)
R.H. Alrefai, P.J. Shiel, L.L. Domier, C.J. D’Arcy, P.H. Berger, S.S. Korban, J. Gen. Virol. 75, 2847–2850 (1994)
D. Guo, E. Maiss, G. Adam, R. Casper, J. Gen. Virol. 76, 1073–1079 (1995)
P.J. Shiel, R.H. Alrefai, L.L. Domier, S.S. Korban, P.H. Berger, Arch. Virol. 140, 1247–1256 (1995)
G.P. Lee, K.H. Ryu, H.R. Kim, C.S. Kim, D.W. Lee, J.S. Kim, Plant Pathol. J. 18, 259–265 (2002)
T. Thokchom, T. Rana, V. Hallan, R. Ram, A.A. Zaidi, Phytoparasitica 37, 375–379 (2009)
M.A. Larkin, G. Blackshields, N.P. Brown, R. Chenna, P.A. McGettigan, H. McWilliam, F. Valentin, I.M. Wallace, A. Wilm, R. Lopez, J.D. Thompson, T.J. Gibson, D.G. Higgins, Bioinformatics 23, 2947–2948 (2007)
D. Martin, P. Lemey, M. Lott, V. Moulton, D. Posada, P. Lefeuvre, Bioinformatics 26, 2462–2463 (2010)
K. Tamura, D. Peterson, N. Peterson, G. Stecher, M. Nei, S. Kumar, Mol. Biol. Evol. 28, 2731–2739 (2011)
P. Librado, J. Rozas, Bioinformatics 25, 1451–1452 (2009)
E.R. Chare, E.S. Holmes, J. Gen. Virol. 85, 3149–3157 (2004)
F. Fonseca, J.D. Neto, V. Martins, G. Nolasco, Arch. Virol. 150, 1607–1619 (2005)
S.S. Krishna, I. Majumdar, N.V. Grishin, Nucleic Acids Res. 31, 532–550 (2003)
S.W. Scott, M.T. Zimmermann, X. Ge, D.J. MacKenzie, Eur. J. Plant Pathol. 104, 155-161 (1998)
Acknowledgments
The authors thank Dr. Donato Boscia from Istituto di Virologia Vegetale CNR, UOS di Bari, Italy, for providing the infected material. This research was supported by S Grant from the Ministry of Education, Youth and Sports of the Czech Republic and Project No. MSM6046070901.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Apple mosaic virus isolates with partial CP sequences. (DOC 64 kb)
Fig. S1
Clusters of 91 ApMV isolates based on the maximum likelihoodphylogenetic tree of 399 nt long central part of CP. Bootstrap values was applied using 1000 replicates. Asterisk (*) marks partial CP sequences. (DOC 151 kb)
Fig. S1
Amino acid alignment of selected isolates. The position of distinctive Gly7 in the zinc-finger motif is highlighted. (PDF 219 kb)
Rights and permissions
About this article
Cite this article
Grimová, L., Winkowska, L., Ryšánek, P. et al. Reflects the coat protein variability of apple mosaic virus host preference?. Virus Genes 47, 119–125 (2013). https://doi.org/10.1007/s11262-013-0925-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-013-0925-z