Abstract
Background
As chronic kidney disease (CKD) progresses, metabolites undergo diverse transformations. Nevertheless, the impact of these metabolic changes on the etiology, progression, and prognosis of CKD remains uncertain. Our objective is to conduct a metabolomics analysis to scrutinize metabolites and identify significant metabolic pathways implicated in CKD progression, thereby pinpointing potential therapeutic targets for CKD management.
Methods
We recruited 145 patients with CKD and determined their mGFR by measuring the plasma iohexol clearance, whereupon we partitioned them into four groups based on their mGFR values. Non-targeted metabolomics analysis was conducted using UPLC-MS/MS assays. Differential metabolites were identified via one-way ANOVA, PCA, PLS-DA, and OPLS-DA analyses employing the MetaboAnalyst 5.0 platform. Ultimately, we performed differential metabolite pathway enrichment analysis, using both the MetaboAnalyst 5.0 platform and the MBRole2.0 database.
Results
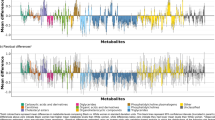
According to the findings of the MBRole2.0 and MetaboAnalyst 5.0 enrichment analysis, six amino acid metabolism pathways were discovered to have significant roles in the progression of CKD, with the glycine, serine, and threonine metabolism pathway being the most prominent. The latter enriched 14 differential metabolites, of which six decreased while two increased concomitantly with renal function deterioration.
Conclusions
The metabolic analysis unveiled that glycine, serine, and threonine metabolism plays a pivotal role in the progression of CKD. Specifically, glycine was found to increase while serine decreased with the deterioration of CKD.





Similar content being viewed by others
Data availability
The original contributions presented in the study are included in the article material, further inquiries can be directed to the corresponding authors.
Code availability
Not applicable.
Abbreviations
- ANOVA:
-
Analysis of variance
- BMI:
-
Body mass index
- CKD:
-
Chronic kidney disease
- eGFR:
-
Estimated glomerular filtration rate
- ESI:
-
Electrospray ionization
- FA:
-
Formic acid
- GFR:
-
Glomerular filtration rate
- HILIC:
-
Hydrophilic interaction chromatography
- IAA:
-
Indole-3-acetic acid
- IS:
-
Indoxyl sulphate
- mGFR:
-
Measured glomerular filtration rate
- 5-MTP:
-
5-Methoxy tryptophan
- m/z :
-
Mass to charge ratio
- NF-κB:
-
Nuclear factor-κB
- OPLS-DA:
-
Orthogonal partial least squares-discriminant analysis
- OSC:
-
Orthogonal signal correction
- PCA:
-
Principal component analysis
- PCS:
-
p-Cresyl sulphate
- PFPA:
-
Perfluoropentanoic acid
- PLS-DA:
-
Partial least squares-discriminant analysis
- RP:
-
Reversed-phase
- TPH-1:
-
Tryptophan hydroxylase-1
- UPLC-MS/MS:
-
Ultra performance liquid chromatograph tandem mass spectrometry
- VIP:
-
Variable importance in the projection
References
Wolf G (2006) Renal injury due to renin-angiotensin-aldosterone system activation of the transforming growth factor-beta pathway. Kidney Int 70(11):1914–1919. https://doi.org/10.1038/sj.ki.5001846
Chasapi SA, Karagkouni E, Kalavrizioti D, Vamvakas S, Zompra A, Takis PG, Goumenos DS, Spyroulias GA (2022) NMR-based metabolomics in differential diagnosis of chronic kidney disease (CKD) subtypes. Metabolites. https://doi.org/10.3390/metabo12060490
Kalim S, Rhee EP (2017) An overview of renal metabolomics. Kidney Int 91(1):61–69. https://doi.org/10.1016/j.kint.2016.08.021
Johnson CH, Ivanisevic J, Siuzdak G (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 17(7):451–459. https://doi.org/10.1038/nrm.2016.25
Hocher B, Adamski J (2017) Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol 13(5):269–284. https://doi.org/10.1038/nrneph.2017.30
Wu G, Zhong J, Chen L, Gu Y, Hong Y, Ma J, Zheng N, Liu AJ, Sheng L, Zhang W, Li H (2020) Effects of the Suxiao Jiuxin pill on acute myocardial infarction assessed by comprehensive metabolomics. Phytomedicine 77:153291. https://doi.org/10.1016/j.phymed.2020.153291
Song Y, Hu T, Gao H, Zhai J, Gong J, Zhang Y, Tao L, Sun J, Li Z, Qu X (2021) Altered metabolic profiles and biomarkers associated with astragaloside IV-mediated protection against cisplatin-induced acute kidney injury in rats: an HPLC-TOF/MS-based untargeted metabolomics study. Biochem Pharmacol 183:114299. https://doi.org/10.1016/j.bcp.2020.114299
Gupta N, Yadav DK, Gautam S, Kumar A, Kumar D, Prasad N (2023) Nuclear magnetic resonance-based metabolomics approach revealed the intervention effect of using complementary and alternative medicine (CAM) by CKD patients. ACS Omega 8(8):7722–7737. https://doi.org/10.1021/acsomega.2c06469
Parker VJ, Fascetti AJ, Klamer BG (2019) Amino acid status in dogs with protein-losing nephropathy. J Vet Intern Med 33(2):680–685. https://doi.org/10.1111/jvim.15436
Rhee EP, Clish CB, Wenger J, Roy J, Elmariah S, Pierce KA, Bullock K, Anderson AH, Gerszten RE, Feldman HI (2016) Metabolomics of chronic kidney disease progression: a case-control analysis in the chronic renal insufficiency cohort study. Am J Nephrol 43(5):366–374. https://doi.org/10.1159/000446484
Chen DQ, Cao G, Chen H, Argyopoulos CP, Yu H, Su W, Chen L, Samuels DC, Zhuang S, Bayliss GP, Zhao S, Yu XY, Vaziri ND, Wang M, Liu D, Mao JR, Ma SX, Zhao J, Zhang Y, Shang YQ, Kang H, Ye F, Cheng XH, Li XR, Zhang L, Meng MX, Guo Y, Zhao YY (2019) Identification of serum metabolites associating with chronic kidney disease progression and anti-fibrotic effect of 5-methoxytryptophan. Nat Commun 10(1):1476. https://doi.org/10.1038/s41467-019-09329-0
Shah VO, Townsend RR, Feldman HI, Pappan KL, Kensicki E, Vander Jagt DL (2013) Plasma metabolomic profiles in different stages of CKD. Clin J Am Soc Nephrol 8(3):363–370. https://doi.org/10.2215/cjn.05540512
Chen YY, Chen DQ, Chen L, Liu JR, Vaziri ND, Guo Y, Zhao YY (2019) Microbiome-metabolome reveals the contribution of gut-kidney axis on kidney disease. J Transl Med 17(1):5. https://doi.org/10.1186/s12967-018-1756-4
Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L, Ge W, Liu W, Liang S, Chen H, Zhang Y, Li J, Xu J, He Z, Chen B, Wang J, Yan H, Zheng Y, Wang D, Zhu J, Kong Z, Kang Z, Liang X, Ding X, Ruan G, Xiang N, Cai X, Gao H, Li L, Li S, Xiao Q, Lu T, Zhu Y, Liu H, Chen H, Guo T (2020) Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182(1):59-72.e15. https://doi.org/10.1016/j.cell.2020.05.032
Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N, Xia J (2022) Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc 17(8):1735–1761. https://doi.org/10.1038/s41596-022-00710-w
López-Ibáñez J, Pazos F, Chagoyen M (2016) MBROLE 2.0-functional enrichment of chemical compounds. Nucleic Acids Res 44(W1):W201-204. https://doi.org/10.1093/nar/gkw253
Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, Raj DS (2016) Role of the gut microbiome in uremia: a potential therapeutic target. Am J Kidney Dis 67(3):483–498. https://doi.org/10.1053/j.ajkd.2015.09.027
Wang X, Yang S, Li S, Zhao L, Hao Y, Qin J, Zhang L, Zhang C, Bian W, Zuo L, Gao X, Zhu B, Lei XG, Gu Z, Cui W, Xu X, Li Z, Zhu B, Li Y, Chen S, Guo H, Zhang H, Sun J, Zhang M, Hui Y, Zhang X, Liu X, Sun B, Wang L, Qiu Q, Zhang Y, Li X, Liu W, Xue R, Wu H, Shao D, Li J, Zhou Y, Li S, Yang R, Pedersen OB, Yu Z, Ehrlich SD, Ren F (2020) Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 69(12):2131–2142. https://doi.org/10.1136/gutjnl-2019-319766
Rysz J, Franczyk B, Ławiński J, Olszewski R, Ciałkowska-Rysz A, Gluba-Brzózka A (2021) The impact of CKD on uremic toxins and gut microbiota. Toxins (Basel). https://doi.org/10.3390/toxins13040252
Barreto FC, Barreto DV, Liabeuf S, Meert N, Glorieux G, Temmar M, Choukroun G, Vanholder R, Massy ZA (2009) Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin J Am Soc Nephrol 4(10):1551–1558. https://doi.org/10.2215/cjn.03980609
Letourneau P, Bataille S, Chauveau P, Fouque D, Koppe L (2020) Source and composition in amino acid of dietary proteins in the primary prevention and treatment of CKD. Nutrients. https://doi.org/10.3390/nu12123892
Kumar MA, Bitla AR, Raju KV, Manohar SM, Kumar VS, Narasimha SR (2012) Branched chain amino acid profile in early chronic kidney disease. Saudi J Kidney Dis Transpl 23(6):1202–1207. https://doi.org/10.4103/1319-2442.103560
Yu Z, Zhai G, Singmann P, He Y, Xu T, Prehn C, Römisch-Margl W, Lattka E, Gieger C, Soranzo N, Heinrich J, Standl M, Thiering E, Mittelstraß K, Wichmann HE, Peters A, Suhre K, Li Y, Adamski J, Spector TD, Illig T, Wang-Sattler R (2012) Human serum metabolic profiles are age dependent. Aging Cell 11(6):960–967. https://doi.org/10.1111/j.1474-9726.2012.00865.x
Hirschel J, Vogel M, Baber R, Garten A, Beuchel C, Dietz Y, Dittrich J, Körner A, Kiess W, Ceglarek U (2020) Relation of whole blood amino acid and acylcarnitine metabolome to age, sex, BMI, puberty, and metabolic markers in children and adolescents. Metabolites. https://doi.org/10.3390/metabo10040149
Wang Y, Zhao M, Wang M, Zhao C (2016) Profiling analysis of amino acids from hyperlipidaemic rats treated with Gynostemma pentaphyllum and atorvastatin. Pharm Biol 54(10):2254–2263. https://doi.org/10.3109/13880209.2016.1152278
Holeček M, Vodeničarovová M (2020) Effects of low and high doses of fenofibrate on protein, amino acid, and energy metabolism in rat. Int J Exp Pathol 101(5):171–182. https://doi.org/10.1111/iep.12368
Irving BA, Carter RE, Soop M, Weymiller A, Syed H, Karakelides H, Bhagra S, Short KR, Tatpati L, Barazzoni R, Nair KS (2015) Effect of insulin sensitizer therapy on amino acids and their metabolites. Metabolism 64(6):720–728. https://doi.org/10.1016/j.metabol.2015.01.008
Yu B, Li AH, Metcalf GA, Muzny DM, Morrison AC, White S, Mosley TH, Gibbs RA, Boerwinkle E (2016) Loss-of-function variants influence the human serum metabolome. Sci Adv 2(8):e1600800. https://doi.org/10.1126/sciadv.1600800
Juhanson P, Kepp K, Org E, Veldre G, Kelgo P, Rosenberg M, Viigimaa M, Laan M (2008) N-acetyltransferase 8, a positional candidate for blood pressure and renal regulation: resequencing, association and in silico study. BMC Med Genet 9:25. https://doi.org/10.1186/1471-2350-9-25
Samynathan R, Subramanian U, Venkidasamy B, Shariati MA, Chung IM, Thiruvengadam M (2022) S-allylcysteine (SAC) exerts renoprotective effects via regulation of TGF-β1/Smad3 pathway mediated matrix remodeling in chronic renal failure. Curr Pharm Des 28(8):661–670. https://doi.org/10.2174/1381612828666220401114301
Stenflo J, Lundwall A, Dahlbäck B (1987) beta-Hydroxyasparagine in domains homologous to the epidermal growth factor precursor in vitamin K-dependent protein S. Proc Natl Acad Sci USA 84(2):368–372. https://doi.org/10.1073/pnas.84.2.368
Manabe S, Marui Y, Ito Y (2003) Total synthesis of mannosyl tryptophan and its derivatives. Chemistry 9(6):1435–1447. https://doi.org/10.1002/chem.200390163
Cheng Y, Li Y, Benkowitz P, Lamina C, Köttgen A, Sekula P (2020) The relationship between blood metabolites of the tryptophan pathway and kidney function: a bidirectional Mendelian randomization analysis. Sci Rep 10(1):12675. https://doi.org/10.1038/s41598-020-69559-x
Fürst P (1989) Amino acid metabolism in uremia. J Am Coll Nutr 8(4):310–323. https://doi.org/10.1080/07315724.1989.10720307
Ruberti B, Machado DP, Vendramini THA, Pedrinelli V, Marchi PH, Jeremias JT, Pontieri CFF, Kogika MM, Brunetto MA (2022) Serum metabolites characterization produced by cats CKD affected, at the 1 and 2 stages, before and after renal diet. Metabolites. https://doi.org/10.3390/metabo13010043
Zeng L, Yu Y, Cai X, Xie S, Chen J, Zhong L, Zhang Y (2019) Differences in serum amino acid phenotypes among patients with diabetic nephropathy, hypertensive nephropathy, and chronic nephritis. Med Sci Monit 25:7235–7242. https://doi.org/10.12659/msm.915735
Hasegawa S, Jao TM, Inagi R (2017) Dietary metabolites and chronic kidney disease. Nutrients. https://doi.org/10.3390/nu9040358
Kim H, Yu B, Li X, Wong KE, Boerwinkle E, Seidelmann SB, Levey AS, Rhee EP, Coresh J, Rebholz CM (2022) Serum metabolomic signatures of plant-based diets and incident chronic kidney disease. Am J Clin Nutr 116(1):151–164. https://doi.org/10.1093/ajcn/nqac054
Li T, Zhang W, Hu E, Sun Z, Li P, Yu Z, Zhu X, Zheng F, Xing Z, Xia Z, He F, Luo J, Tang T, Wang Y (2021) Integrated metabolomics and network pharmacology to reveal the mechanisms of hydroxysafflor yellow A against acute traumatic brain injury. Comput Struct Biotechnol J 19:1002–1013. https://doi.org/10.1016/j.csbj.2021.01.033
Funding
This study was supported by grants from the National Natural Science Foundation of China (Grant No. 81873631, 81370866, 81070612) and Science and Technology Plan Project of Guangzhou (Grant No. 202002020047, 202007040003), The Science and Technology Development Fund, Macau SAR (File no. 0032/2018/A1).
Author information
Authors and Affiliations
Contributions
All authors have read and approved the manuscript. JK and XG: Conceptualization, methodology, and writing—original draft preparation. XL and HP: Funding acquisition. YD and CA: Visualization and Supervision. JL and TsT: Investigation. LT and TT: Data curation and Validation. XL and HP: Writing-review, funding, and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
The studies involving human participants were reviewed and approved by the Institutional Review Board of Ethics Commission of Kiang Wu Hospital (KWH 2018-001). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kang, J., Guo, X., Peng, H. et al. Metabolic implications of amino acid metabolites in chronic kidney disease progression: a metabolomics analysis using OPLS-DA and MBRole2.0 database. Int Urol Nephrol 56, 1173–1184 (2024). https://doi.org/10.1007/s11255-023-03779-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11255-023-03779-8