Abstract
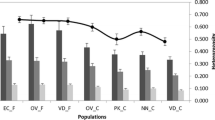
Designing strategies for conservation and improvement livestock should be based on assessment of genetic characteristics of populations under consideration. In Oman, conservation programs for local livestock breeds have been started. The current study assessed the genetic diversity and conservation potential of local chickens from Oman. Twenty-nine microsatellite markers were analyzed in 158 birds from six agroecological zones: Batinah, Dhofar, North Hajar, East Hajar, Musandam, and East Coast. Overall, a total of 217 alleles were observed. Across populations, the average number of alleles per locus was 7.48 and ranged from 2 (MCW98 and MCW103) to 20 (LEI094). The mean expected heterozygosity (H E) was 0.62. Average fixation index among populations (F ST) was 0.034, indicating low population differentiation, while the mean global deficit of heterozygotes across populations (F IT) was 0.159. Based on Nei’s genetic distance, a neighbor-joining tree was constructed for the populations, which clearly identified the Dhofar population as the most distant one of the Omani chicken populations. The analysis of conservation priorities identified Dhofar and Musandam populations as the ones that largely contribute to the maximal genetic diversity of the Omani chicken gene pool.

Similar content being viewed by others
References
Al-Saadi, N., 2013. The Animal and Plant Genetic Resources Center; summary of the proposed strategy. In: SQU (ed), Proceedings of the Symposium on biotechnology and conservation of species from arid regions, 10-13/2//2013, Muscat, Oman.
Berthouly, C., Bed’Hom, B., Tixier-Boichard, M., Chen, C.F., Lee, Y.P., Laloë, D., Legros, H., Verrier, E., and Rognon, X., 2008. Using molecular markers and multivariate methods to study the genetic diversity of local European and Asian chicken breeds. Animal Genetics, 39, 121-129
Bodzsar, N., Eding, H., Revay, T., Hidas, A., and Weigend, S., 2009. Genetic diversity of Hungarian indigenous chicken breeds based on microsatellite markers. Animal Genetics, 4, 516–523
Boettcher, P.J., Tixier-Boichard, M., Toro, M.A., Simianer, H., Eding, H., Gandini, G., Joost, S., Garcia, D., Colli, D., Ajmone-Marsan, P., and GLOBALDIV Consortium, 2010. Objectives, criteria and methods for using molecular genetic data in priority setting for conservation of animal genetic resources. Animal Genetics, 41, 64–77
Caballero, A., and Toro, M.A., 2002. Analysis of genetic diversity for the management of conserved subdivided populations. Conservation Genetics, 3, 289-299
Caballero, A., Rodríguez-Ramilo, S.T., Ávila, V., and Fernández, J., 2010. Management of genetic diversity of subdivided populations in conservation programmes. Conservation Genetics, 11, 409-419
Chen, G.H., Bao, W.B., Shu, J., Ji, C.L., Wang, M.Q., Eding, H., Muchadeyi, F., and Weigend, S., 2008. Assessment of population structure and genetic diversity of 15 Chinese indigenous chicken breeds using microsatellite markers. Asian-Australasian Journal of Animal Sciences, 21, 331-339
Cuc, N.T., Simianer, H., Eding, H., Tieu, H.V., Cuong, V.C., Wollny, C.B., Groeneveld, L.F., and Weigend, S., 2010. Assessing genetic diversity of Vietnamese local chicken breeds using microsatellites. Animal Genetics, 41, 545-547
DGMAN (Directorate General of Meteorology and Air Navigation), 2012. Annual report of weather forecast for Oman. Muscat, Oman
FAO, 2004. The state of the world’s animal genetic resources for food and agriculture. In: B. Rischkowsky and D. Pilling (eds), Rome, Italy.
FAO, 2010. Chicken genetic resources used in smallholder production systems and opportunities for their development. In: P. Sørensen (ed), FAO Smallholder Poultry Production, Rome, Italy
Felsenstein, J., 1995. PHYLIP (Phylogeny Inference Package). 1995, Department of Genetics, University of Washington, Seattle, USA
Gandini, G.C., and Oldenbroek, J.K., 1999. Choosing the conservation strategy. In: J.K. Oldenbroek (ed), Genebanks and the conservation of farm animal genetic resources, 11–31. DLO, Netherlands.
Granevitze, Z., Hillel, J., Feldman, M., Six, A., Eding, H., and Weigend, S., 2009. Genetic structure of a wide-spectrum chicken gene pool. Animal Genetics, 40, 686-693
Halima, H.M., 2007. Phenotypic and genetic characterization of indigenous chicken populations in Northwest Ethiopia. (PhD thesis, University of the Free State).
Jesuyon, O.M.A., and Salako, A.E., 2013. Variability and predictability of productive and body traits of Fulani ecotype chicken. African Journal of Agricultural Research, 8, 6178-6184
Kadim, I., Al-Marzooqi, W., Al-Waheebi, S., and Mahgoub, O., 2009. A project for improving egg and meat production of Omani local chicken through genetic and phenotypic selection programmes (Annual Project Report).
Kirkpatrick, S., Gelatt, C., and Vecchi, M.P., 1983. Optimization by simulated annealing. Science, 220, 671–680
Leroy, G., Kayang, B.B., Youssao, I.A., Yapi-Gnaore’, C.V., Osei-Amponsah, R., Loukou, N.E., Fotsa, J.C., Benabdeljelil, K., Bed’hom, B., Tixier-Boichard, M., and Rognon, X., 2012. Gene diversity, agroecological structure and introgression patterns among village chicken populations across North, West and Central Africa. BMC Genetics, 13, 34
MAF (Ministry of Agriculture and Fisheries), 2011. Agriculture and livestock, five-year research strategy (2011–2015). Muscat, Oman
Mahgoub, O., 2012. Characterization, evaluation and conservation of indigenous animal genetic resources in the Sultanate of Oman, Project progress report. Sultan Qaboos University, Muscat, Oman.
Mtileni, B.J., Muchadeyi, F.C., Maiwashe, A., Groeneveld, E., Dzama, K., and Weigend, S., 2011. Genetic diversity and conservation of South African indigenous chicken populations. Journal of Animal Breeding and Genetics, 128, 209-218
Mtileni, B.J., Muchadeyi, F.C., Maiwashe, A., Chimonyo, M., and Dzama, K., 2012. Conservation and utilization of indigenous chicken genetic resources in Southern Africa. World’s Poultry Science Journal, 68, 727-748
Muchadeyi, F.C., Eding, H., Wollny, C.B., Groeneveld, E., Makuza, S.M., Shamseldin, R., Simianer, H., and Weigend, S., 2007. Absence of population substructuring in Zimbabwe chicken ecotypes inferred using microsatellite analysis. Animal Genetics, 38, 332-339
Park, S.D., 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. (PhD thesis, University of Dublin).
Peakall, R., and Smouse, P.E., 2006. GENALEX 6: genetic analysis in excel population, genetic software for teaching and research. Molecular Ecology Notes, 6, 288-295
Pérez-Figueroa, A., Saura, M., Fernández, J., Toro, M.A., and Caballero, A., 2009. METAPOP—a software for the management and analysis of subdivided populations in conservation programs. Conservation Genetics, 10, 1097–1099
Raymond, M., and Rousset, F., 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86, 248-249
Weigend, S., Groenen, M.A., Tixier-Boichard, M., Vignal, A., Hillel, J., Wimmers, K., Burke, T., and Tanila, M.A., 1998. Development of strategy and application of molecular tools to assess biodiversity in chicken genetic resources (AVIANDIV) EC contract No. BIO4-CT98-0342.
Acknowledgments
We are grateful to the farmers who participated in our study and thank the Agricultural Directorates and Research Centers in Oman for coordinating field visits. We highly appreciate the technical assistance of Mrs. Annett Weigend, Mr. Maik Przyklenk, and Mrs. Natalie Janus at the Institute of Farm Animal Genetics, Mariensee. This research was embedded in a project aiming at the genetic characterization of Omani livestock resources, funded by the Sultan Qaboos University through HM Fund SR/AGR/ANVS/08/01.
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Oman map showing the geographical distribution of the six agroecological zones (in circles) and sampling areas within each zone (triangles). See material and methods section for population name abbreviations. (DOC 131 kb)
Table S1
Loci names, number of alleles, observed (HO) and expected (HE) heterozygosity calculated for each microsatellite across 6 Omani local chicken populations. (DOC 48 kb)
Appendix 1: parameters and formulas used for conservation analyses
Appendix 1: parameters and formulas used for conservation analyses
To assess conservation priorities, two methods were used as described by Caballero and Toro (2002). For the first method, for each population i, the global genetic diversity GDtli is calculated excluding this population, then the loss of genetic diversity (L GD ) is calculated as follows:
where GDt is the total genetic diversity, GDtli the global genetic diversity after removing population i, and L GD the loss of genetic diversity.
Therefore, the higher the contribution of a population to the global genetic diversity is, the higher will be the decrease of genetic diversity when that population is excluded from the analysis. Negative values indicate that global genetic diversity is increased when that population is excluded.
For the second method, the contribution that a population i will add to a pool with maximum genetic diversity (GDpool) could be achieved by obtaining the values of c i that maximize GDpool as follows:
where c i is restricted to c ≥ 0 and ∑ n i = 1 c i = 1 with n being the number of populations, f ii the coancestry coefficient within population i, and D ij Nei’s minimum distance between population i and j (Caballero and Toro 2002).
Rights and permissions
About this article
Cite this article
Al-Qamashoui, B., Simianer, H., Kadim, I. et al. Assessment of genetic diversity and conservation priority of Omani local chickens using microsatellite markers. Trop Anim Health Prod 46, 747–752 (2014). https://doi.org/10.1007/s11250-014-0558-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-014-0558-9