Abstract
One of the most powerful strategies to investigate biology we have as scientists, is the ability to transfer genetic material in a controlled and deliberate manner between organisms. When applied to livestock, applications worthy of commercial venture can be devised. Although initial methods used to generate transgenic livestock resulted in random transgene insertion, the development of SCNT technology enabled homologous recombination gene targeting strategies to be used in livestock. Much has been accomplished using this approach. However, now we have the ability to change a specific base in the genome without leaving any other DNA mark, with no need for a transgene. With the advent of the genome editors this is now possible and like other significant technological leaps, the result is an even greater diversity of possible applications. Indeed, in merely 5 years, these ‘molecular scissors’ have enabled the production of more than 300 differently edited pigs, cattle, sheep and goats. The advent of genome editors has brought genetic engineering of livestock to a position where industry, the public and politicians are all eager to see real use of genetically engineered livestock to address societal needs. Since the first transgenic livestock reported just over three decades ago the field of livestock biotechnology has come a long way—but the most exciting period is just starting.
Similar content being viewed by others
Introduction
Biology has many facets and our ability to utilize novel biological pathways increases every year. As scientists we strive to develop tools and strategies to help us tease apart biological process so we can better understand them. To achieve this understanding, biologists often turn to the powerful approach involving gene transfer enabling the consequence of alterations in gene activity to be studied in vivo. Since the first steps in the early 1970s involving the transformation of bacteria, the successful transfer of genes into first mammals and then plants quickly followed, with similar progress in fish and insects soon achieved. The first transgenic livestock announced in the mid-1980s (Hammer et al. 1985) followed the pioneering work of Palmiter and Brinster in mice (Brinster et al. 1981). Since then we have come a long way, with an explosion of activity in recent years.
The field of genetically engineered livestock has been driven by technological advances. This 30 year journey started slowly, with pronuclear injection (PNI) being the first tool in the kit (Hammer et al. 1985). Although conceptually simple—delivering DNA by injection through a fine glass needle into one of the pronuclei of a fertilised egg—this method is technically demanding and those who could successfully accomplish it were given great respect by the research community. PNI was king for the first decade of transgenic livestock research, with the commercial enterprises emerging on the back of this technique focused on producing human biomedical proteins in animal bioreactors (Jänne and Alhonen 1998; Kind and Schnieke 2008). But it has limitations. Efficiency of generating founder animals was low, and the injected DNA construct integrated randomly into the genome resulting in unpredictable transgene expression profiles. The field needed to progress.
Just over 10 years ago, oncoretroviruses were first employed to produce transgenic livestock (Chan et al. 1998; Cabot et al. 2001). While harnessing the innate ability of these replication defective viral vectors to transduce the livestock zygote dramatically increased the efficiency with which founder transgenic animals could be created, it quickly became obvious that there were issues with silencing of transgene expression in subsequent generations. The move from oncoretroviruses such as MoMLV to lentiviruses increased the transgenesis efficiencies further; Whitelaw et al. (2004) used an EIAV-based virus encoding eGFP to produce 40 founder piglets, 37 of which were transgenic and 35 of which expressed eGFP. Breeding from a subset of these for four generations revealed no loss of gene expression as assessed by visual GFP fluorescence and western blot for GFP protein (Whitelaw et al. unpublished data), in contrast to other studies with lentivirus transgenes where gene silencing was observed as assessed by loss of GFP fluorescence and DNA methylation (Hofmann et al. 2006). This impressive efficiency has allowed lentiviruses to be used to create a cohort of transgenic founder animals to model human disease (Kostic et al. 2013). However, viral vectors remain limited to a small cargo carrying capacity (around 8 Kb for lentiviruses) and the fact that they can only act as unidirectional delivery vehicles. Offering comparable efficiencies, transposon-based transgenesis (Carlson et al. 2011; Jakobsen et al. 2011) is less constrained with regard to vector design and the vagaries of transgene silencing (Ivics et al. 2014).
Gene targeting by homologous recombination
Those working on transgenic livestock looked around to see what was being achieved in other mammalian species. In particular attention was drawn to what was technically possible in rodent research, where in addition to random transgene integration through pronuclear injection, the ability to perform gene targeting was possible. Gene targeting is made possible through homologous recombination (HR) which involves the exchange of nucleotides between two similar or identical DNA sequences (Capecchi 1989). In this way a gene can be targeted for disruption, termed knockout (KO), or used as a docking site for transgene insertion, termed knock in (KI)—this achievement attracted the first of two Nobel Prizes in this field.
HR in mammalian cells is an inefficient process, so relied on inclusion of a selectable marker in the construct to enable only the cells containing gene targeted events to survive. Subsequently more elegant strategies involving recombination-steps, driven for example by Cre recombinase (Nagy et al. 2009), enabled removal of the undesired marker gene. The latter has also enabled targeting of a transgene to a given genetic ‘harbour’, usually intended as a site permissive for expression of the transgene (Bronson et al. 1996; Wallace et al. 2000). The multiple steps involved and the low targeting efficiencies achieved meant that HR was not practical in zygotes; a cell-based system was required and the mouse research community had a very good one. Embryonic stem cells (ESCs), derived from the preimplantation embryo, have the ability to both self-renew and retain pluripotential characteristics (Torres-Padilla and Chambers 2014; Martello and Smith 2014). The first property facilitates the lengthy process of gene targeting while the latter allows the engineered cell to contribute to the germline after transfer into the early embryo (Capecchi 1989).
HR and ESCs transformed mouse-based research in the 1980s and until recently formed the mainstay for this research community, enabling the development of huge and important research resources. The race was on to achieve similar progress in livestock. But there was a hurdle. Livestock ESCs had not and still have not been isolated (Malaver-Ortega et al. 2012). For reasons that remain a mystery no robust livestock ESCs have ever been demonstrated, even though huge advances in our understanding of both rodent and human ESC biology (Torres-Padilla and Chambers 2014) and the requirements for their maintenance in culture has been achieved (Buehr et al. 2008). An alternative method of producing ESC-like cells through a process involving regression of differentiated cells was developed in 2006 (Takahashi and Yamanaka 2006). These induced pluripotent stem cells (iPSC) can self-renew and like ESCs can differentiate into many different cells types including the germline but are not derived from the early embryo. Again this development was pioneered in mice—winning the second Nobel Prize in the field—and was quickly transferred into human biology. However, while there have been some limited successes in livestock, iPSCs remain surprisingly difficult to isolate and even harder to maintain (Telugu et al. 2011; Ezashi et al. 2012; Nowak-Imialek and Niemann 2012).
Cloning first enabled gene targeting in livestock
Since robust pluripotent livestock cells have to-date proven beyond reach, the livestock research community had to come up with an alternative—which they did in the mid-1990s. That alternative manifested in the birth of Dolly the sheep (Wilmut et al. 1997). Next year will be the 20th anniversary of Dolly and in these intervening years considerable use of somatic cell nuclear transfer (SCNT), more commonly referred to as cloning, has been the method of choice for many teams engaged with producing transgenic livestock (Kues and Niemann 2011; Prather 2013; Wolf et al. 2014). SCNT utilises primary cells grown in culture. During this in vitro phase manipulation of the donor genome utilising methodologies based on HR can be applied, with the selected transgenic cell then used to reconstitute an enucleated oocyte. The resulting transgenic animals are clones, derived from genetically identical parental cells. SCNT has enabled transgenic livestock research to develop since the late 1990s. Nevertheless, even with technical advances to simplify the technique such as handmade cloning (Peura and Vajta 2003), SCNT remains technically difficult and only a few labs around the world have truly mastered it.
By the turn of the millennium we were able to do gene addition, gene KO and gene KI—the latter two only by SCNT—in livestock. Given the cost involved in these studies, the need for SCNT and the complexity of HR driven gene targeting, transgenic livestock research has focussed primarily on biomedical applications. Here notable progress has been made in the extent of resources now available for xenotransplantation applications (Klymiuk et al. 2010; Satyananda et al. 2013; Cooper et al. 2014) and the commercial success of animal bioreactor derived products (Bösze et al. 2008). Agricultural applications, however, have lagged behind primarily due to concerns over public acceptance of livestock containing transgenes in the food chain.
The revolution that is genome editors
1996 was a watershed year for the generation of engineered animals; not only did it include the birth of Dolly but also the generation of the first programmable nuclease (Kim et al. 1996). The former was trumpeted loudly by the media with reverberations still echoing around this field. It was over a decade for the latter to emerge as the technical revolution it is. Nearly 20 years on we now have a variety of tools and techniques at our disposal for the generation of engineered livestock species. Until recently we have only been able to dream of the ability to change a specific base in the genome without leaving any other DNA footprint; or the ability to induce precise insertions or deletions easily and efficiently in the germline of livestock. With the advent of the genome editors this is now possible.
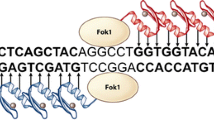
Designer nucleases are used to generate a double strand break (DSB) at a desired genomic locus and can be divided into two broad categories; synthetic and natural. The first category includes the original zinc finger nuclease (ZFN; Kim et al. 1996) and the newer transcription activator like effector nuclease (TALEN; Christian et al. 2010) both of which are modular proteins containing an adaptable DNA binding domain fused to the nuclease domain of FokI. In the case of ZFNs each individual zinc finger binds three DNA bases whereas each TAL repeat binds a single base. Both ZFNs and TALENs are employed as pairs which recognise opposing DNA strands and orientate such that their fused FokI monomers are brought together on the intervening sequence to form an active enzyme dimer that cleaves both strands. In a refinement of this system the FokI monomers have been mutated such that heterodimerisation is obligate for FokI cutting (Miller et al. 2007; Doyon et al. 2011).
The second category includes meganucleases (Smith et al. 2006) and the newest, and currently the most popular of the designer nucleases, the clustered regularly interspaced short palindromic repeat/CRISPR associated gene (CRISPR/Cas) system (Cong et al. 2013; Mali et al. 2013; Jinek et al. 2013). Uptake of meganucleases by the livestock research community has not been widespread, presumably due to the laborious protein re-design and optimisation that is required to repurpose these molecules to a novel DNA sequence (Smith et al. 2006), however, effort to develop this nuclease tool continues (Ménoret et al. 2013). In contrast, the CRISPR/Cas system, which was first described just over 2 years ago, has seen an unprecedented exponential increase in its use (Seruggia and Montoliu 2014). This relatively simple system is adapted from an innate immune mechanism common to many bacteria and archaea, the function of which is to protect against invading viruses. The most widely used system at present is based on the CRISPR/Cas9 of Streptococcus pyogenes and involves a short guide RNA (sgRNA) sequence complexed with Cas9 nuclease. Specificity is determined by hybridisation between the 20 ribonucleotides of the complexed Cas9/guide and the nascent DNA target sequence, further restricted to sites immediately proximal to a protospacer adjacent motif (PAM) sequence (Cong et al. 2013; Mali et al. 2013; Jinek et al. 2013).
Following generation of a DSB at the desired locus, repair can occur in one of two ways; non-homologous end joining (NHEJ) or homology dependent repair (HDR; Fig. 1). In most cases of DSBs are repaired by NHEJ, with the two ends of the break being brought together and ligated. As a consequence of endogenous nuclease activity at the cut site this process is error prone and often results in the introduction small insertions/deletions (indels) at the repair site (Kanaar et al. 1998). Alternatively, if a repair template is provided in trans, evoking HDR in addition to NHEJ, the introduction of desired changes to the sequence at the targeted locus can be achieved (Kanaar et al. 1998). Deletion of regions of the genome can be achieved by generating of a pair of DSBs flanking the region to be deleted and their subsequent repair by NHEJ (Carlson et al. 2012; He et al. 2015b; Whitworth et al. 2014; Fig. 2).
Routes to genome edited livestock. Designer nucleases have been successfully used to modify both zygotes and somatic cells. Modification and selection of fibroblasts coupled with SCNT has resulted in the generation of HDR and NHEJ edited livestock. NHEJ edited animals have been produced via zygote CPI whereas, to date, HDR edited animals have not been reported from edited zygotes
The utility of double strand breaks generated by genome editors. A cartoon depiction of the double strand break (DSB) repair mechanisms. Non homologous end joining (NHEJ) is an error prone process that re-joins the end of the DSB, often resulting in small insertions/deletions (blue) and subsequent gene disruption. Homology dependent repair is a faithful process that uses a homologous template to repair the DSB. Providing a repair template, either as a single stranded oligonucleotide or double stranded DNA, allows specific modifications (green) to be introduced to the genome. Creation of simultaneous DSBs flanking a region of the genome can result in deletion of the intervening sequence (yellow) and repair of the DSBs by either NHEJ or HDR. (Color figure online)
Nickases are modified nucleases that only cut one strand of DNA, and mutagenesis of both FokI and Cas9 has resulted in nickase versions of these enzymes. By designing reagents such that staggered nicks are created at the target site (e.g. 2 pairs of ZFNickases), a DSB still occurs (Kim et al. 2012) and is repaired by either NHEJ or HDR. One potential advantage of this approach is that binding at off-target sites results in a nick rather than a DSB, with subsequent repair by the break excision repair (BER) pathway which leaves no mark on the genome. It is too early to know how useful the nickases strategy will be given the reduced efficiency associated with this Cas9 variant but the theoretical promise of reduced off-targets seems to be real (Ren et al. 2014; Frock et al. 2015).
So much achieved in such a short period of time
In the last 5 years, genome editors have been used to mediate the generation of more than 300 edited pigs, cattle, sheep and goats (Table 1). These animals can potentially serve as organ donors (Hauschild et al. 2011; Li et al. 2015), disease models (Tan et al. 2013), bioreactors (Liu et al. 2013) or founder animals of genetic lines with enhanced productivity (Proudfoot et al. 2015) or disease resistance traits (Lillico et al. 2013; Wu et al. 2015). To achieve this, our community has been progressively expanding the livestock genome engineering toolbox to include state-of-the-art technologies, first ZFNs (Yang et al. 2011; Whyte et al. 2011), then TALENs (Carlson et al. 2012) and CRISPR/Cas9 (Hai et al. 2014; Fig. 3).
The creation of the first genome edited animals relied on the modification of primary cells which were then used as nuclear donors for embryo reconstruction in SCNT (Hauschild et al. 2011; Carlson et al. 2012; Fig. 3). An efficient alternative, direct modification of zygotes by cytoplasmic injection (CPI) of the editors, soon followed (Fig. 4a; Lillico et al. 2013; Hai et al. 2014) rekindling the microinjection skills used for the first transgenic livestock (Fig. 3). Adding to these initial reports that described NHEJ events, the ability to introduce defined sequences into a targeted locus through HDR, using either single strand DNA oligonucleotides (ssODN; Tan et al. 2013) or plasmids as repair templates (Liu et al. 2013; Wu et al. 2015) has been demonstrated for livestock. Rather than depending on random changes at the target site introduced by the error prone NHEJ repair pathway, these defined sequence changes allow either more precise gene knockout or targeted integration of various transgenes, and importantly make allele swapping possible (Fig. 2).
SCNT has been the primary method to deliver nuclease mediated genetic changes into livestock. To date, 33 out of 43 reported successes utilise SCNT and resulted in 267 edited live animals (Table 1). This focus on SCNT reflects the lead position this technology has had for the last couple of decades in livestock biology, especially for pigs, since one can apply nucleases then pre-select editing events in vitro. At time of writing, SCNT is the only published way to create livestock with defined changes by HDR (Fig. 3).
The combination of genome editors and SCNT has proven to be powerful. It is possible to obtain cells with bi-allelic modifications in one-step by marker-free dilution cloning (Tan et al. 2013), or if necessary FACS sorting (Whyte et al. 2011; Reyes et al. 2014) or drug selection (Liu et al. 2013; Wu et al. 2015). Moreover, simultaneous targeting of different genes has allowed bi-allelic modification of up to three genes at the same time (Reyes et al. 2014; Li et al. 2015). Cloning using such cells has resulted in an average 76 % editing rate in live born pigs (Table 2); some of these animals contain gene inactivating indels resulting from NHEJ (Lutz et al. 2013; Whitworth et al. 2014) or ssODN mediated HDR (Tan et al. 2013), while others have site-specific insertions of transgenes by HR (Kwon et al. 2013; Wu et al. 2015; Cui et al. 2015).
Production of editor modified animals via SCNT is hugely successful, but remains tied to the drawbacks associated with cloning. In the published reports using editors and SCNT, cloning efficiency has been low, being only 1.2 % (278/23,216) for pigs and 0.6 % (58/10,510) for other livestock (Table 2). On average, production of one edited live pig requires reconstruction of 130 embryos, which would be challenging without ready access to abattoir-sourced oocytes. The cloning efficiency is partly affected by donor cell quality: prolonged culture and multiple manipulations of the cells decrease their efficiency as nuclear donors. Because of this, some studies have required re-cloning to obtain more founder animals (Hauschild et al. 2011), especially when several genes were targeted simultaneously (Lutz et al. 2013; Reyes et al. 2014). In addition, SCNT (even in the absence of nuclease treatment) is often associated with problems such as birth defects, abortions and early postnatal death (Keefer 2015).
To circumvent issues associated with cloning, some research groups have adopted direct microinjection of editing reagents to the cytoplasm of zygotes. This approach has been effective in generating edited livestock animals using all three designer nucleases systems in pigs (Lillico et al. 2013; Hai et al. 2014; Whitworth et al. 2014; Wang et al. 2015), cattle (Proudfoot et al. 2015) and sheep (Han et al. 2014; Proudfoot et al. 2015). Although application of CPI has not been as widely used as SCNT in livestock genome editing (41 edited animals by CPI vs 267 animals by SCNT (Table 1), its use is gaining momentum. This shift presumably reflects the simplicity and versatility of CPI over SCNT. While the editing efficiency in live-born animals is lower for CPI (in pigs 37 % for CPI vs 76 % for SCNT), reflecting the lack of selection that takes place during the in vitro phase of SCNT, CPI only requires an average of 24 embryos to produce one edited pig, a five-fold improvement on that currently reported for SCNT (Table 2). Furthermore, one of the biggest advantages of CPI is that it can be applied to zygotes from any desired parental cross, maintaining genetic diversity in progeny. By contrast, SCNT tends to use genetic material from a clonal cell population, resulting in offspring that are genetically identical to the donor cells and thus requiring subsequent outcrossing to maintain genetic variation. SCNT, however, enables selection of a specific mutation prior to production of animals and potentially enables access to genetic lines where the correct embryo donors are not readily available. Thus, both CPI and SCNT offer the opportunity to utilise the range of editing events that are produced at a given locus.
Given that there is scope for improving the proportion of live-born edited animals following CPI, we anticipate that the numbers of both donor and recipient animals required per edited offspring will continue to be reduced. Combined with freedom from cloning related problems and greater choice in genetic background, CPI may prove to be the more compelling method—at least for agricultural applications. In the meantime there are two areas that require further investigation; founder mosaicism and efficient HDR. Mosaicism is commonly observed in edited animals produced by CPI (Lillico et al. 2013; Han et al. 2014; Proudfoot et al. 2015), and while this could be problematic with respect to analysis of phenotype in the founder generation, our group routinely breed CPI generated F0 pigs (homozygous, heterozygous or mosaic) to produce F1 offspring with the desired genotype (Fig. 4b; unpublished data). Other groups have confirmed germline transmission from mosaics by germ cell genotyping (Hai et al. 2014; Wang et al. 2015). Ideally mosaicism following CPI needs to be reduced; options currently being explored include maximizing editor concentration while controlling toxicity and off-target mutations or alternatively delivering Cas9 as protein rather than mRNA. HDR mediated editing via CPI has yet to be reported in live-born animals. However, given the high efficiency of multiplexing editing events in rodents (Wang et al. 2013; Yang et al. 2013), the recent success by co-injecting editors and ssODNs into in vitro cultured bovine embryos (Wei et al. 2015), and data from our own lab indicating that allele swap animals can be produced in zygotes (unpublished results), this is unlikely to prove a significant limitation.
Live genome edited pigs produced by TALEN injection into zygotes. a Founder NHEJ animals born 2012 (Lillico et al. 2013). b Third generation piglets derived from NHEJ founder animals
Expanding current trends
Although most of the initial goals in livestock genetic engineering focused on agriculture (Pursel et al. 1989), the combination of rudimentary (although at the time innovative) tools, insufficient knowledge of the genetic consequence of modifying the initial targets (e.g. growth hormone: Pursel et al. 1989) and lack of public support lead to a dearth of funding, both public and commercial, in this area. The result was the development of applications on the biomedical-biotechnological interface: animal bioreactors, xenotransplantation and, more recently, large animal models of human disease (Jänne and Alhonen 1998; Kind and Schnieke 2008; Cooper et al. 2014; Kostic et al. 2013). Given this spectrum of applications, impacts of genome editing may be quite variable. For animal bioreactor projects genome editing technology can enable refined expression strategies and possibly be used to augment post-translational effects. While editing technology could accelerate the development of even more animal resources for xenotransplantation, it is perhaps at the interface of these two applications that genome editing technology may come to the fore. The remarkable report of chimeric mice that carried a rat cell derived pancreas (Kobayashi et al. 2010) reignited enthusiasm for the possibility of animals producing human organs, or at least human cells. The passage of time since the original report may have dampened some of the initial optimism, but effort in this direction continues. Success would be spectacular.
Within the biomedical arena there is a growing realisation that small animal models, although delivering great mechanistic insight into disease in the model organism, are insufficient as translational tools for converting this knowledge from bench research to bedside applications. In many situations we would benefit from the use of larger animals to model both the development of disease pathology and the testing of intervention strategies. Genome editing enables the mutation of endogenous livestock gene homologues of known causative or associated human disease loci. Although there are no projects yet that have fully exploited this new technology, there are already a growing number of projects based on the ‘older’ transgenic technologies that have demonstrated that this belief in large animal disease models is justified (Wolf et al. 2014; Aigner et al. 2010).
A path to agriculture
Man has pursued the selective breeding of animals for a long time. Initially our Mesolithic ancestors identified animals for their ability to breed in captivity based on aspects of temperament and social structure. Now engineering enables the targeting introduction of mutations providing increased genetic variation for the animal breeder to utilise. Although some agricultural applications of GM have been pursued by the research community, industry has been shy of engaging with the traditional transgenic technologies. That reluctance to directly engage with projects involving livestock appears to have eased with the advent of genome editing technology.
Historically animal breeding relied on selection by individual farmers of breeding stock with visibly desirable traits. Bioinformatic use of genetics allows a more refined selection process but still relies on selection of randomly segregating loci that are predicted to underlie advantageous traits (Daetwyler et al. 2013; Van Eenennaam et al. 2014; Hill 2014). Genome editing enables introgression of single genetic loci in contrast to current breeding regimes. It enables access to inaccessible variation: variation that doesn’t exist in a given breeding population (Lillico et al. 2013), i.e. variation out-with the breeding gene pool (a currently discussed scenario is introgression of the polled trait into elite Holstein cattle). It also offers solutions to non-segregation of beneficial and undesirable traits due to physical proximity of the underlying loci in the genome. Alternatively, genome editing offers a route to eliminate deleterious alleles from a gene pool.
Genome editing could also be used to modify quantitative traits that are affected by many loci (associated with a number of quantitative trait nucleotides). In the natural state low levels of recombination will limit the rate favourable alleles will arise together in selected individuals. This presents challenges for breeding regimes aiming to improve such a quantitative trait. Simulations show that even relatively modest multiplexing of genome editing targets has great potential for increasing the response to selection in breeding programmes over improvement by genomic selection alone (Jenko et al. 2015).
Many sectors of the animal agriculture industry, including those working with pigs, cattle, and chickens are now actively engaged with the technology through collaborations with the academic community. There remain the perceived barriers to adoption of these technologies in animal agriculture including public opinion and the regulatory environment; but these factors are in flux. Public opinion is often largely influenced by a vocal minority of concerned members of our society, but within society there also exists support and interest in the potential for genetically engineered livestock to make a contribution to the global challenge of food security. This is increasingly being reflected by the constructive media portrayal of this biotechnology. In turn, the political mood is also changing; for example, the headline in the UK press early 2015 “EU regulation on GMOs not ‘fit for purpose’ say UK MPs”. Nevertheless, at time of writing, we still see the regulatory approval for arguably the world’s lead GM product (the AquAdvantage salmon) in the quagmire that occurs when politics overrides the scientific evidence-based position and the approved regulatory process (Van Eenennaam and Muir 2011).
It is hoped that genome edited livestock (Fahrenkrug et al. 2010), with their lack of introduced transgenes, will find a smoother path through the regulatory system. In plants the regulatory bodies view this enzyme-based technology as they would chemical or radiation induced mutagenesis; the latter two methods have a long history of unregulated use but lack the specificity afforded by genome editing tools. We believe that in this regulatory environments, and based on the specificity and ‘footprintless’ nature of genome editing, gene editing animals will successfully navigate both the political and regulatory landscape.
What is next for genome engineering
Transgenic livestock were first produced in the mid-1980s and subsequently through the use of HR and SCNT, gene targeting in large animals has been possible since the late 1990s (Clark and Whitelaw 2003). Gene editing brings nothing conceptually new to the table. Rather, this set of tools greatly facilitates what was already possible with traditional methods, increasing the rate at which new projects can be delivered. This is fast becoming reality with an incredible pace of progress being evident—for those engaged with the literature there is the real feeling that as many new lines of engineered large animals have been produced in the last few years as in the previous three decades.
Some believe genome editing tools provide the best imaginable technology for mutating the germline. Indeed it is hard at the moment to imagine what could be better. Nevertheless there are remaining challenges. We need to improve efficiency of editing within a given population of cells (destined for SCNT) and in the zygote and overcome mosaicism. In our work with zygotes we regularly achieve 30 % editing frequency with delivery of editors—ZFN, TALEN and CRISPR/Cas9—to the cytoplasm of livestock. We should aspire to at least >50 % and why not frequencies approaching or even achieving 100 %.
We need to further refine our predictive editor design algorithms. The scope to expand the repertoire of editing reagents continues through the development of Cas9 variants (Kleinstiver et al. 2015; Ran et al. 2015) and meganucleases (Ménoret et al. 2013) is already materializing with the promise that many more will be forthcoming. For real utility in addressing multi-quantitative nucleotides underlying a quantitative locus we will need the ability to multiplex editing events. Conceptually this could be challenging, given the possibility for multiple target sites in a given genome to undergo inter-site events, resulting in deletion or other forms of recombination, yet the production mice with of three consecutively edited sites has been reported.
For reasons both practical and public perception based, the concern about off-targets must be addressed. Off-target effects occur because the editing complex relies on base-recognition affinity for targeting but can cut at a lower frequency at similar non-target sites (analogous to the star activity exhibited by restriction endonucleases). Although a much discussed point, the emerging evidence now suggest that off-targets may be rare events in mice (Iyer et al. 2015), supporting previous human cell data (Kim et al. 2015). Nevertheless there is probably still room for improvement, although the debate about what could be tolerated for a given application remains to be resolved. To address this aspect a multitude of strategies are being evaluated. For example, masking Cas9 with a fusion peptide preventing activity until cleaved by a small molecule (Davis et al. 2015), expanding the TALEN RDV repertoire (Miller et al. 2015), dimerisation of the editing enzyme (Wright et al. 2015; Zetsche et al. 2015), use of nickases which cause single strand-breaks rather than double-strand breaks therefore evoking different DNA repair processes (Frock et al. 2015) and, further in this vein, the inhibition of NHEJ (Maruyama et al. 2015; Chu et al. 2015).
Continued development of genome editing tools will accelerate livestock biotechnology through their ease of use. Where it took several painfully taxing years for several groups around the world to produce alpha-1,3 galactosyltransferase null pigs, this can now rapidly and easily be achieved through the use of genome editors. This ‘catch-up’ phenomenon is not unique to these tools and reflects all aspects of technological advance (most obvious in our ability to sequence genomes). And like other significant technology leaps, this results is more and greater diversity in applications. Concerns of off-targets are reducing and the political landscape increasingly supportive. Although currently there is a crowded intellectual property landscape enveloping the genome editors (e.g. Sherkow 2015), paths through this legal environment will resolve with time. We are only at the start of this wave of advance—the world for livestock biotechnology is about to get very exciting.
References
Aigner B, Renner S, Kessler B, Klymiuk N, Kurome M, Wünsch A, Wolf E (2010) Transgenic pigs as models for translational biomedical research. J Mol Med 88(7):653–664
Bao L, Chen H, Jong U, Rim C, Li W, Lin X, Huang H (2014) Generation of GGTA1 biallelic knockout pigs via zinc-finger nucleases and somatic cell nuclear transfer. Sci China Life Sci 57(2):263–268
Bösze Z, Baranyi M, Bruce C, Whitelaw A (2008) Producing recombinant human milk proteins in the milk of livestock species. In: Bioactive components of milk. Springer, pp 357–395
Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD (1981) Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell 27(1):223–231
Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O (1996) Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci 93(17):9067–9072
Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, Smith A (2008) Capture of authentic embryonic stem cells from rat blastocysts. Cell 135(7):1287–1298
Cabot RA, Kuhholzer B, Chan AW, Lai L, Park KW, Chong KY, Prather RS (2001) Transgenic pigs produced using in vitro matured oocytes infected with a retroviral vector. Anim Biotechnol 12(2):205–214. doi:10.1081/ABIO-100108347
Capecchi MR (1989) Altering the genome by homologous recombination. Science 244(4910):1288–1292
Carlson DF, Garbe JR, Tan W, Martin MJ, Dobrinsky JR, Hackett PB, Fahrenkrug SC (2011) Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Res 20(5):1125–1137
Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Fahrenkrug SC (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci 109(43):17382–17387
Chan AW, Homan EJ, Ballou LU, Burns JC, Bremel RD (1998) Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proc Natl Acad Sci USA 95(24):14028–14033
Chen F, Wang Y, Yuan Y, Zhang W, Ren Z, Jin Y et al (2015) Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genom 42:437–444
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186(2):757–761
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R (2015) Increasing the efficiency of homology-directed repair for CRISPR–Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33(5):543–548
Clark J, Whitelaw B (2003) A future for transgenic livestock. Nat Rev Genet 4(10):825–833
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Marraffini LA (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823
Cooper DK, Satyananda V, Ekser B, Windt DJ, Hara H, Ezzelarab MB, Schuurman HJ (2014) Progress in pig-to-non-human primate transplantation models (1998–2013): a comprehensive review of the literature. Xenotransplantation 21(5):397–419
Cui C, Song Y, Liu J, Ge H, Li Q, Huang H et al (2015) Gene targeting by TALEN-induced homologous recombination in goats directs production of β-lactoglobulin-free, high-human lactoferrin milk. Sci Rep 5:10482
Daetwyler HD, Calus MP, Pong-Wong R, de los Campos G, Hickey JM (2013) Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193(2):347–365
Davis KM, Pattanayak V, Thompson DB, Zuris JA, Liu DR (2015) Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat Chem Biol 11(5):316–318
Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Holmes MC (2011) Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods 8(1):74–79
Ezashi T, Telugu B, Roberts R (2012) Induced pluripotent stem cells from pigs and other ungulate species: an alternative to embryonic stem cells? Reprod Domest Anim 47(s4):92–97
Fahrenkrug S, Blake A, Carlson D, Doran T, Van Eenennaam A, Faber D, Li N (2010) Precision genetics for complex objectives in animal agriculture. J Anim Sci 88(7):2530–2539
Frock RL, Hu J, Meyers RM, Ho Y-J, Kii E, Alt FW (2015) Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33(2):179–186
Hai T, Teng F, Guo R, Li W, Zhou Q (2014) One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res 24(3):372
Hammer RE, Pursel VG, Rexroad CE Jr, Wall RJ, Bolt DJ, Ebert KM, Brinster RL (1985) Production of transgenic rabbits, sheep and pigs by microinjection. Nature 315(6021):680–683
Han H, Ma Y, Wang T, Lian L, Tian X, Hu R, Li N (2014) One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Front Agric Sci Eng 1(1):2–5
Hauschild J, Petersen B, Santiago Y, Queisser A-L, Carnwath JW, Lucas-Hahn A, Schwinzer R (2011) Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci 108(29):12013–12017
He J, Li Q, Fang S, Guo Y, Liu T, Ye J, Hu X (2015a) PKD1 mono-allelic knockout is sufficient to trigger renal cystogenesis in a mini-pig model. Int J Biol Sci 11(4):361
He Z, Proudfoot C, Mileham AJ, McLaren DG, Whitelaw CBA, Lillico SG (2015b) Highly efficient targeted chromosome deletions using CRISPR/Cas9. Biotechnol Bioeng 112(5):1060–1064
Hill WG (2014) Applications of population genetics to animal breeding, from Wright, Fisher and Lush to genomic prediction. Genetics 196(1):1–16
Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, Pfeifer A (2006) Epigenetic regulation of lentiviral transgene vectors in a large animal model. Mol Ther 13(1):59–66
Huang J, Guo X, Fan N, Song J, Zhao B, Ouyang Z, Yi X (2014) RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol 193(3):1496–1503
Ivics Z, Garrels W, Mátés L, Yau TY, Bashir S, Zidek V, Rülicke T (2014) Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons. Nat Protoc 9(4):810–827
Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, Skarnes WC (2015) Off-target mutations are rare in Cas9-modified mice. Nat Methods 12(6):479
Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, Mikkelsen JG (2011) Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res 20(3):533–545. doi:10.1007/s11248-010-9438-x
Jänne J, Alhonen L (1998) Transgenic livestock as bioreactors. In: Mammary gland transgenesis: therapeutic protein production, vol 161
Jenko J, Gorjanc G, Cleveland MA, Varshney RK, Whitelaw CBA, Woolliams JA, Hickey JM (2015) Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs. Genet Sel Evol 47(1):1–14
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. Elife 2:e00471
Kanaar R, Hoeijmakers JH, van Gent DC (1998) Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol 8(12):483–489
Keefer CL (2015) Artificial cloning in domestic animals Proc. Natl Acad Sci 112:8874–8878
Kim Y-G, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93(3):1156–1160
Kim E, Kim S, Kim DH, Choi B-S, Choi I-Y, Kim J-S (2012) Precision genome engineering with programmable DNA-nicking enzymes. Genome Res 22(7):1327–1333
Kim D, Bae S, Park J, Kim E, Kim S, Yu HR et al (2015) Digenome-seq: genome-wide profiling of CRISPR–Cas9 off-target effects in human cells. Nat Methods 12:237–243
Kind A, Schnieke A (2008) Animal pharming, two decades on. Transgenic Res 17(6):1025–1033
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z et al (2015) Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523:481–485
Klymiuk N, Aigner B, Brem G, Wolf E (2010) Genetic modification of pigs as organ donors for xenotransplantation. Mol Reprod Dev 77(3):209–221
Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Knisely A (2010) Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell 142(5):787–799
Kostic C, Lillico SG, Crippa SV, Grandchamp N, Pilet H, Philippe S, Sarkis C (2013) Rapid cohort generation and analysis of disease spectrum of large animal model of cone dystrophy. PLoS One 8(8):e71363
Kues WA, Niemann H (2011) Advances in farm animal transgenesis. Prev Vet Med 102(2):146–156
Kwon D-N, Lee K, Kang M-J, Choi Y-J, Park C, Whyte JJ et al (2013) Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep 3:1981
Lee K, Kwon D-N, Ezashi T, Choi Y-J, Park C, Ericsson AC, Walters EM (2014) Engraftment of human iPS cells and allogeneic porcine cells into pigs with inactivated RAG2 and accompanying severe combined immunodeficiency. Proc Natl Acad Sci 111(20):7260–7265
Li P, Estrada JL, Burlak C, Tector AJ (2013) Biallelic knockout of the α-1, 3 galactosyltransferase gene in porcine liver-derived cells using zinc finger nucleases. J Surg Res 181(1):e39–e45
Li F, Li Y, Liu H, Zhang H, Liu C, Zhang X, Du Y (2014) Production of GHR double-allelic knockout Bama pig by TALENs and handmade cloning. Yi chuan=Hereditas/Zhongguo yi chuan xue hui bian ji 36(9):903–911
Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, Downey SM (2015) Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation 22(1):20–31
Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C et al (2013) Live pigs produced from genome edited zygotes. Sci Rep 3:2847
Liu X, Wang Y, Guo W, Chang B, Liu J, Guo Z et al (2013) Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat Commun 4
Liu X, Wang Y, Tian Y, Yu Y, Gao M, Hu G, Guo Z (2014) Generation of mastitis resistance in cows by targeting human lysozyme gene to β-casein locus using zinc-finger nucleases. Proc R Soc Lond B Biol Sci 281(1780):20133368
Luo J, Song Z, Yu S, Cui D, Wang B, Ding F et al (2014) Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PloS One 9:e95225
Lutz AJ, Li P, Estrada JL, Sidner RA, Chihara RK, Downey SM, Ivary B (2013) Double knockout pigs deficient in N-glycolylneuraminic acid and Galactose α-1, 3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation 20(1):27–35
Malaver-Ortega LF, Sumer H, Liu J, Verma PJ (2012) The state of the art for pluripotent stem cells derivation in domestic ungulates. Theriogenology 78(8):1749–1762
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826
Martello G, Smith A (2014) The nature of embryonic stem cells. Annu Rev Cell Dev Biol 30(1):647
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL (2015) Increasing the efficiency of precise genome editing with CRISPR–Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol 33(5):538–542
Ménoret S, Fontanière S, Jantz D, Tesson L, Thinard R, Rémy S, Anegon I (2013) Generation of Rag1-knockout immunodeficient rats and mice using engineered meganucleases. FASEB J 27(2):703–711
Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L, Rupniewski I, Kim KA (2007) An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25(7):778–785
Miller JC, Zhang L, Xia DF, Campo JJ, Ankoudinova IV, Guschin DY, Lam SC (2015) Improved specificity of TALE-based genome editing using an expanded RVD repertoire. Nat Methods 12(5):465–471
Nagy A, Mar L, Watts G (2009) Creation and use of a cre recombinase transgenic database. In: Wurst W, Kühn R (eds) Gene knockout protocols, 2nd edn. Humana Press, Totowa, pp 365–378
Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M et al (2014) Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE 9:e106718
Nowak-Imialek M, Niemann H (2012) Pluripotent cells in farm animals: state of the art and future perspectives. Reprod Fertil Dev 25(1):103–128
Peura TT, Vajta G (2003) A comparison of established and new approaches in ovine and bovine nuclear transfer. Cloning Stem Cells 5(4):257–277
Prather RS (2013) Pig genomics for biomedicine. Nat Biotechnol 31(2):122–124
Proudfoot C, Carlson DF, Huddart R, Long CR, Pryor JH, King TJ, Whitelaw CBA (2015) Genome edited sheep and cattle. Transgenic Res 24(1):147–153
Pursel VG, Pinkert CA, Miller KF, Bolt DJ, Campbell RG, Palmiter RD, Hammer RE (1989) Genetic engineering of livestock. Science 244(4910):1281–1288
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Makarova KS (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520(7546):186–191
Ren X, Yang Z, Mao D, Chang Z, Qiao H-H, Wang X, Liu L-P (2014) Performance of the Cas9 nickase system in Drosophila melanogaster. G3: Genes| Genomes|. Genetics 4(10):1955–1962
Reyes LM, Estrada JL, Wang ZY, Blosser RJ, Smith RF, Sidner RA, Miner AC (2014) Creating class I MHC–null pigs using guide RNA and the Cas9 endonuclease. J Immunol 193(11):5751–5757
Satyananda V, Hara H, Ezzelarab MB, Phelps C, Ayares D, Cooper DK (2013) New concepts of immune modulation in xenotransplantation. Transplantation 96(11):937
Seruggia D, Montoliu L (2014) The new CRISPR–Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals. Transgenic Res 23(5):707–716
Sherkow JS (2015) Law, history and lessons in the CRISPR patent conflict. Nat Biotechnol 33(3):256–257
Smith J, Grizot S, Arnould S, Duclert A, Epinat J-C, Chames P, Bravo J (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 34(22):e149–e149
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC (2013) Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci 110(41):16526–16531
Telugu BPV, Ezashi T, Sinha S, Alexenko AP, Spate L, Prather RS, Roberts RM (2011) Leukemia inhibitory factor (LIF)-dependent, pluripotent stem cells established from inner cell mass of porcine embryos. J Biol Chem 286(33):28948–28953
Torres-Padilla M-E, Chambers I (2014) Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development 141(11):2173–2181
Van Eenennaam AL, Muir WM (2011) Transgenic salmon: a final leap to the grocery shelf? Nat Biotech 29(8):706–710
Van Eenennaam AL, Weigel KA, Young AE, Cleveland MA, Dekkers JC (2014) Applied animal genomics: results from the field. Annu Rev Anim Biosci 2(1):105–139
Wallace H, Ansell R, Clark J, McWhir J (2000) Pre-selection of integration sites imparts repeatable transgene expression. Nucleic Acids Res 28(6):1455–1464
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153(4):910–918. doi:10.1016/j.cell.2013.04.025
Wang Y, Du Y, Shen B, Zhou X, Li J, Liu Y et al (2015) Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep 5:8256
Watanabe M, Nakano K, Matsunari H, Matsuda T, Maehara M, Kanai T, Kuramoto M (2013) Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS One 8(10):e76478
Wei J, Wagner S, Lu D, Maclean P, Carlson DF, Fahrenkrug SC et al (2015) Efficient introgression of allelic variants by embryo-mediated editing of the bovine genome. Sci Rep 5:11735
Whitelaw CBA, Radcliffe PA, Ritchie WA, Carlisle A, Ellard FM, Pena RN, Mitrophanous KA (2004) Efficient generation of transgenic pigs using equine infectious anaemia virus (EIAV) derived vector. FEBS Lett 571(1):233–236
Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Walters EM (2014) Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod 114:121723
Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, Prather RS (2011) Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev 78(1):2
Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KHS (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385(6619):810–813
Wolf E, Braun-Reichhart C, Streckel E, Renner S (2014) Genetically engineered pig models for diabetes research. Transgenic Res 23(1):27–38
Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA (2015) Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci 112(10):2984–2989
Wu H, Wang Y, Zhang Y, Yang M, Lv J, Liu J, Zhang Y (2015) TALE nickase-mediated SP110 knockin endows cattle with increased resistance to tuberculosis. Proc Natl Acad Sci 112(13):E1530–E1539
Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Yang Y (2013) Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS One 8(12):e84250
Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Tian J (2011) Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res 21(6):979
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154(6):1370–1379. doi:10.1016/j.cell.2013.08.022
Yao J, Huang J, Hai T, Wang X, Qin G, Zhang H et al (2014) Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci Rep 4:6926
Yu S, Luo J, Song Z, Ding F, Dai Y, Li N (2011) Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 21(11):1638
Zetsche B, Volz SE, Zhang F (2015) A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat Biotechnol 33(2):139–142
Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Lai S (2014) Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci 72(6):1175–1184
Acknowledgments
This research team is supported by the BBSRC through ISPG and BB/L007371/1, InnovateUK through Catalyst 131782 and CLDC 102085, and by CHDI and Genus plc. We are a member of the EU COST Action SALAAM BM1308.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Tan, W., Proudfoot, C., Lillico, S.G. et al. Gene targeting, genome editing: from Dolly to editors. Transgenic Res 25, 273–287 (2016). https://doi.org/10.1007/s11248-016-9932-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-016-9932-x