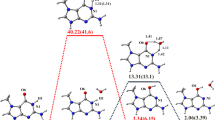
Abstract
Methylated nucleosides play an important role in DNA/RNA function, and may affect republication by interrupting the base-pairing and base-stacking. In order to investigate the effect of methylation on the interaction between nucleic acid bases, this work presents the hydrogen-bonding and stacking interactions between 5-methylcytosine and guanine (G), cytosine (C) and G, 1-methyladenine and thymine (T), as well as adenine and T. Geometry optimization and potential energy surface scan have been performed for the involved complexes by MP2 calculations. The interaction energies, which were corrected for the basis-set superposition error by the full Boys–Bernardi counterpoise correction scheme, were used to evaluate the interaction intensity of these nucleic acid bases. The atoms in molecules theory and natural bond orbital analysis have been performed to study the hydrogen bonds in these complexes. The result shows that the methyl substitute contributes the stability to these complexes because it enhances either the hydrogen bonding or the staking interaction between nucleic acid bases studied.






Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Colot V, Rossignol JL (1999) Eukaryotic DNA methylation as an evolutionary device. BioEssays 21:402–411
Squires JE, Preiss T (2010) Function and detection of 5-methylcytosine in eukaryotic RNA. Epigenomics 2:709–715
Hill G, Paukku Y (2011) Theoretical determination of one-electron redox potentials for DNA bases, base pairs, and stacks. J Phys Chem A 115:4804–4810
Falnes PO, Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J (2004) AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell 16:107–116
Luisi B, Orozco M, Šponer J, Luque FJ, Shakked Z (1998) On the potential role of the amino nitrogen atom as a hydrogen bond acceptor in macromolecules. J Mol Biol 279:1123–1129
Krüger T, Bruhn C, Steinborn D (2004) Synthesis of [(MeCyt)2H]I—structure and stability of a dimeric threefold hydrogen-bonded 1-methylcytosinium 1-methylcytosine cation. Org Biomol Chem 2:2513–2516
Ramakrishnan B, Sundaralingam M (1995) Crystal structure of the A-DNA decamer d(CCIGGCCm5CGG) at 1.6 angstroms showing the unexpected wobble I.m5C base pair. Biophys J 69:553–559
Gao W-l, Li D, Xiao Z-x, Liao Q-p, Yang H-x, Li Y-x, Ji L, Wang Y-l (2011) Detection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placentas. Hypertens Res 34:655–661
Zhu X, Liang J, Li F, Yang Y, Xiang L, Xu J (2011) Analysis of associations between the patterns of global DNA hypomethylation and expression of DNA methyltransferase in patients with systemic lupus erythematosus. Int J Dermatol 50:697–704
Katiyar SK, Nandakumar V, Vaid M (2011) (–)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16(INK4a), by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis 32:537–544
Watson JD, Crick FHC (1953) A structure for deoxyribose nucleic acid. Nature 171:731–738
Parthasarathi R, Subramanian V (2006) Hydrogen bonding of DNA base pairs and information entropy: from molecular electron density perspective. Chem Phys Lett 418:530–534
Dkhissi A, Blossey R (2007) Performance of DFT/MPWB1K for stacking and H-bonding interactions. Chem Phys Lett 439:35–39
Thiviyanathan V, Somasunderam A, Volk DE, Hazra TK, Mitra S, Gorenstein DG (2008) Base-pairing properties of the oxidized cytosine derivative, 5-hydroxy uracil. Biochem Biophys Res Commun 366:752–757
Padermshoke A, Katsumoto Y, Masaki R, Aida M (2008) Thermally induced double proton transfer in GG and wobble GT base pairs: a possible origin of the mutagenic guanine. Chem Phys Lett 457:232–236
Yewski P (2010) Structural and energetic consequences of oxidation of d(ApGpGpGpTpT) telomere repeat unit in complex with TRF1 protein. J Mol Struct (Theochem) 16:1797–1807
Villani G (2006) Theoretical investigation of hydrogen transfer mechanism in the guanine–cytosine base pair. Chem Phys 324:438–446
Meng FC, Xu WR, Liu CB (2004) Theoretical study of various H-bond nonclassical A:T and G:C pairs. J Mol Struct (Theochem) 677:85–90
Atacharyya D, Koripella SC, Mitra A, Rajendran VB, Sinha B (2007) Theoretical analysis of noncanonical base pairing interactions in RNA molecules. J Biosci 32:809–813
Ailov VI, Anisimov VM, Kurita N, Hovorun D (2005) MP2 and DFT studies of the DNA rare base pairs: the molecular mechanism of the spontaneous substitution mutations conditioned by tautomerism of bases. Chem Phy Lett 412:285–293
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian03, revision. Gaussian, Inc. Wallingford CT
Hobza P, Šponer J (2002) Toward true DNA base-stacking energies: MP2, CCSD(T), and complete basis set calculations. J Am Chem Soc 124:11802–11808
Hobza P, Zahradnik R (1988) Intermolecular complexes. Elsevier, Amsterdam
van Duijneveldt FB, van Duijneveldt-van de Rijdt JGCM, van Lenthe JH (1994) State of the art in counterpoise theory. Chem Rev 94:1873–1885
Carpenter JE, Weinhold F (1988) Analysis of the geometry of the hydroxymethyl radical by the “different hybrids for different spins” natural bond orbital procedure. J Mol Struct (Theochem) 169:41–62
Foster JP, Weinhold F (1980) Natural hybrid orbitals. J Am Chem Soc 102:7211–7218
Alkorta I, Elguero J, Foces–Foces C (1996) Dihydrogen bonds (A–H···H–B). Chem Commun 14:1633–1637
Alkorta I, Rozas I, Elguero J (1998) Bond length–electron density relationships: from covalent bonds to hydrogen bond interactions. Struct Chem 9:243–247
Bader RFW (1991) A quantum theory of molecular structure and its applications. Chem Rev 91:893–901
Bader RFW (1998) Bifurcated hydrogen bonds: three-centered interactions. J Phys Chem A 102:9925–9932
Jurecka P, Hobza P (2003) True stabilization energies for the optimal planar hydrogen-bonded and stacked structures of guanine···cytosine, adenine···thymine, and their 9- and 1-methyl derivatives: complete basis set calculations at the MP2 and CCSD(T) levels and comparison with experiment. J Am Chem Soc 125:15608–15613
Desiraju GR (1991) The C–H.cntdot.cntdot.cntdot.O hydrogen bond in crystals: what is it? Acc Chem Res 24:290–296
Starikov EB, Steiner T (1997) Computational support for the suggested contribution of C–HO=C interactions to the stability of nucleic acid base pairs. Acta Cryst 53:345–347
Och U, Popelier PLA (1995) Characterization of C–H–O hydrogen bonds on the basis of the charge density. J Phys Chem 99:9747–9754
Acknowledgments
Financial supports from the National Natural Science Foundation of China (20875038), the Research Program of State Key Laboratory of Food Science and Technology (SKLF-ZZB-201207), and the Program of Innovation Team of Jiangnan University (2008CXTD01) are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, Q., Qiu, Z., Wang, H. et al. The effect of methylation on the hydrogen-bonding and stacking interaction of nucleic acid bases. Struct Chem 24, 55–65 (2013). https://doi.org/10.1007/s11224-012-0027-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-012-0027-x