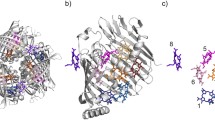
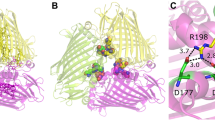
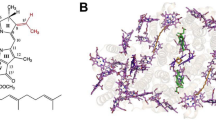
Abstract
The absorbance spectrum of the Fenna-Matthews-Olson protein—a component of the antenna system of Green Sulfur Bacteria—is always one of two types, depending on the species of the source organism. The FMO from Prosthecochloris aestuarii 2K has a spectrum of type 1 while that from Chlorobaculum tepidum is of type 2. The previously reported crystal structures for these two proteins did not disclose any rationale that would explain their spectral differences. We have collected a 1.3 Å X-ray diffraction dataset of the FMO from Prosthecochloris aestuarii 2K, which has allowed us to identify an additional Bacteriochlorophyll-a molecule with chemical attachments to both sides of the central magnesium atom. A new analysis of the previously published X-ray data for the Chlorobaculum tepidum FMO shows the presence of a Bacteriochlorophyll-a molecule in an equivalent location but with a chemical attachment from only one side. This difference in binding is shown to be predictive of the spectral type of the FMO.





Similar content being viewed by others
Abbreviations
- Bchl-a :
-
Bacteriochlorophyll-a
- Cbl :
-
Chlorobaculum
- FMO:
-
Fenna–Matthews–Olson Protein
- PDB:
-
Protein Data Bank
- Pel :
-
Pelodictyon
- Ptc :
-
Prosthecochloris
- r.m.s.:
-
Root mean square
References
Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Mariarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D58:1948–1954
Bairoch A, Boeckmann B, Ferro S, Gasteiger E (2004) Swiss-Prot: juggling between evolution and stability. Brief Bioinform 5:39–55. doi:10.1093/bib/5.1.39
BenShem A, Frolow F, Nelson N (2004) Evolution of photosystem I—from symmetry through pseudosymmetry to asymmetry. FEBS Lett 564:274–280. doi:10.1016/S0014-5793(04)00360-6
Berman HM, Westbrook J, Feng Z, Gilliand G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235
Blankenship RE, Matsuura K (2003) Antenna complexes from green photosynthetic bacteria. In: Green BR, Parson WW (eds) Light-harvesting antennas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 195–217
Bricogne G (1997) The Bayesian statistical viewpoint on structure determination: basic concepts and examples. In: Carter CW, Sweet RM (eds) Methods in enzymology 276A. New York, Academic Press, pp 361–423
Brixner T, Stenger J, Vaswani HM, Minhaeng C, Blankenship RE, Fleming GR (2005) Two-dimensional spectroscopy of electronic couplings in photosynthesis. Nature 434:625–628. doi:10.1038/nature03429
Camara-Artigas A, Blankenship RE, Allen JP (2003) The structure of the FMO protein from Chlorobium tepidum at 2.2 Å resolution. Photosynth Res 75:49–55. doi:10.1023/A:1022406703110
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi:10.1107/S0907444904019158
Engh RA, Huber R (1991) Accurate bond and angle parameter for X-ray protein structure refinement. Acta Crystallogr A47:392–400
Engle GS, Calhoun TR, Read EL, Ahn T-K, Mančal T, Cheng Y-C, Blankenship RE, Fleming GR (2007) Evidence for wavelike energy transfer through quantum coherence in photosynthetic systems. Nature 446:782–786. doi:10.1038/nature05678
Fenna RE, Ten Eyck LF, Matthews BW (1977) Atomic coordinates for the chlorophyll core of a bacteriochlorophyll a-protein from green photosynthetic bacteria. Biochem Biophys Res Commun 75:751–755. doi:10.1016/0006-291X(77)91536-4
Fiedor L (2006) Hexacoordination of bacteriochlorophyll in photosynthetic antenna LH1. Biochemistry 45:1910–1918. doi:10.1021/bi0514055
Francke C, Amesz J (1997) Isolation and pigment composition of the antenna system of four species of green sulphur bacteria. Photosynth Res 52:137–146. doi:10.1023/A:1005845828676
Imhoff JF (2003) Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int J Syst Evol Microbiol 53:941–951. doi:10.1099/ijs.0.02403-0
Louwe RJW, Vrieze J, Aartsma TJ, Hoff AJ (1997a) Toward an integral interpretation of the optical steady-state spectra of the FMO-complex of Prosthecochloris aestuarii. 1. An investigation with linear-dichroic absorbance-detected magnetic resonance. J Phys Chem B 101(51):11273–11279. doi:10.1021/jp972215+
Louwe RJW, Vrieze J, Hoff AJ, Aartsma TJ (1997b) Toward an integral interpretation of the optical steady-state spectra of the FMO-complex of Prosthecochloris aestuarii. 2. Exciton simulations. J Phys Chem B 101(51):11280–11287. doi:10.1021/jp9722162
Lovell SC, Davis IW, Arendall WBIII, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by C-alpha geometry: phi, psi, and C-beta deviation. Proteins 50:437–450. doi:10.1002/prot.10286
McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi:10.1107/S0907444905001617
Melkozernov AN, Olson JM, Li YF, Allen JP, Blankenship RE (1998) Orientation and excitonic interactions of the Fenna–Matthews–Olson Protein in membranes of the green sulfur bacterium Chlorobium tepidum. Photosynth Res 56:315–328. doi:10.1023/A:1006082513522
Müh F, Madjet ME-A, Adolphs J, Abdurahman A, Rabenstein B, Ishikita H, Knapp E-W, Renger T (2007) α-Helices direct excitation energy flow in the Fenna–Matthews–Olson protein. Proc Natl Acad Sci USA 140:16862–16867. doi:10.1073/pnas.0708222104
O’Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C (2004) 3DCoffee: combining protein sequences and structures within multiple sequence alignments. J Mol Biol 340:385–395. doi:10.1016/j.jmb.2004.04.058
Olson JM (2004) The FMO protein. Photosynth Res 80:181–187. doi:10.1023/B:PRES.0000030428.36950.43
Olson JM, Ke B, Thompson KH (1974) Exciton interaction among chlorophyll molecules in bacteriochlorophyll a protein and bacteriochlorophyll a reaction center complexes from green bacteria. Biochim Biophys Acta 430:524–537 Errata (1976). 440, 763
Otwinowski ZW, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM (eds) Methods in Enzymology, 276A. Academic Press, New York, pp 307–326
Pearlstein RM (1992) Theory of the optical spectra of the bacteriochlorophyll a antenna protein trimer from Prosthecochloris aestuarii. Photosynth Res 31:213–226. doi:10.1007/BF00035538
Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6:458–463. doi:10.1038/8263
Savikhin S, Buck DR, Struve WS (1997) Pump-probe anisotropies of Fenna–Matthews–Olson protein trimers from Chlorobium tepidum: a diagnostic for exciton localization? Biophys J 73:2090–2096. doi:10.1016/S0006-3495(97)78239-0
Savikhin S, Buck DR, Struve WS (1998) Toward level-to-level energy transfers in photosynthesis: the Fenna–Matthews–Olson protein. J Phys Chem B 102:5556–5565. doi:10.1021/jp981186f
Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64:112–122. doi:10.1107/S0108767307043930
Shibata Y, Saga Y, Tamiaki H, Itoh S (2007) Polarized fluorescence of aggregated bacteriochlorophyll c and baseplate bacteriochlorophyll a in single chlorosomes isolated from Chloroflexus aurantiacus. Biochemistry 46:7062–7068. doi:10.1021/bi0623072
Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung L-W, Read RJ, Adams PD (2008) Iterative model building, structure refinement, and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. doi:10.1107/S090744490705024X
Tronrud DE, Matthews BW (1993) Refinement of the structure of a water-soluble antenna complex from green photosynthetic bacteria by incorporation of the chemically determined amino acid sequence. In: Deisenhofer J, Norris JR (eds) The photosynthetic reaction center, vol 1. Academic Press, Inc., San Diego
Tronrud DE, Schmid MF, Matthews BW (1986) Structure and X-ray amino acid sequence of a bacteriochlorophyll a protein from Prosthecochloris aestuarii refined at 1.9 Å resolution. J Mol Biol 188:443–454. doi:10.1016/0022-2836(86)90167-1
van Mourik F, Verwijst RR, Mulder JM, van Grondelle R (1994) Singlet-triplet spectroscopy of the light-harvesting BChl a complex of Prosthecochloris aestuarii. The nature of the low-energy 825 nm transition. J Phys Chem 98(40):10307–10312. doi:10.1021/j100091a054
Vulto SIE, de Baat MA, Louwe RJW, Permentier HP, Neef T, Miller M, van Amerongen H, Aartsma TJ (1998a) Exciton simulations of optical spectra of the FMO complex from the green sulphur bacterium Chlorobium tepidum at 6 K. J Phys Chem B 102(47):9577–9582. doi:10.1021/jp982095l
Vulto SIE, Neerken S, Louwe RJW, de Baat MA, Amesz J, Aartsma TJ (1998b) Excited-state structure and dynamics in FMO antenna complexes from photosynthetic green sulphur bacteria. J Phys Chem B 102(51):10630–10635. doi:10.1021/jp983003v
Wen J, Zhang H, Gross ML, Blankenship RE (2009) Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc Natl Acad Sci USA 106(15):6134–31639. doi:10.1073/pnas.0901691106
Wendling M, Przyjalgowski MA, Gülen D, Vulto SIE, Aartsma TJ, van Grondelle R, van Amerongen H (2002) The quantitative relationship between structure and polarized spectroscopy in the FMO comples of Prosthecochloris aestuarii: refining experiments and simulations. Photosynth Res 71:99–123. doi:10.1023/A:1014947732165
Whitten WB, Olson JM, Pearlstein RM (1980) Seven-fold exciton splitting of the 810-nm band in bacteriochlorophyll a-proteins from green photosynthetic bacteria. Biochim Biophys Acta 591:203–207. doi:10.1016/0005-2728(80)90234-0
Acknowledgments
The authors would like to thank a number of individuals for their assistance and advice in performing this work. Roger Fenna provided advice on the purification and crystallization of this protein that was helpful in recreating the crystal form he had used in his earlier work. George Sheldrick answered a great many questions about the operation of his program ShelxL. Tom Terwilliger helped out with the operation of his Phenix.AutoBuild program. Tom Womack, Clemens Vonrhein, and others at Global Phasing, Ltd helped with the operation of AutoBuster. Anthony Addlagatta provided advice and assistance in crystal handling and data collection. X-ray diffraction data were collected at ALS beamline 8.2.2. Figures 1, 2 and 3 were created using the program PyMOL (http://www.pymol.org/). R. E. Blankenship gratefully acknowledges support from grant #DE-FG02-07ER15846 from the Energy Biosciences program of the Basic Energy Sciences division of the US Department of Energy. D. E. Tronrud wishes to thank B. W. Matthews for agreeing to host this project in his lab.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tronrud, D.E., Wen, J., Gay, L. et al. The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth Res 100, 79–87 (2009). https://doi.org/10.1007/s11120-009-9430-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-009-9430-6