Abstract
Aims
We tested whether chemical content of individual Tansy plants influences aboveground arthropod and belowground soil microbial community composition.
Methods
We use Tansy chemical defence composition as focus for our analysis, given that changes in chemical defence are a dominant mechanism expected to modify aboveground and belowground community composition and nutrient cycling. Detailed assessment of arthropod food webs were made on tansy chemotypes. Next generation soil DNA analyses were used to assess soil bacterial community.
Results
Tansy plants defended by Camphor and Borneol had higher total soil bacterial diversity than control plants (soils under non-Tansy plants) and Tansy chemotypes containing high concentrations of Thujon. Furthermore, the frequency of bacterial genera with a significant role in litter mineralization processes decreased with time in soil associated with Camphor and Borneol chemotypes, indicative of complete microbial decomposition and mineralization of plant material. No such changes occurred in soils associated with Thujon chemotypes. Thujon plants had fewer aphids, ladybirds, spiders, and Orius sp. than other chemotypes.
Conclusions
Our experiments revealed that Thujon chemotypes had different associated aboveground arthropod and belowground soil microbial communities than Camphor or Borneol chemotypes. The common qualitative response between aboveground and belowground communities to Tansy plant defense is consistent with our hypothesis and extensive literature on plant defense and aboveground and belowground feedbacks.
Similar content being viewed by others
Introduction
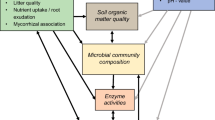
Understanding what controls the structure and functioning of terrestrial ecosystems has been greatly enhanced by considering aboveground and belowground food chains as coupled systems (Wardle et al. 2004; Moore et al. 2004; Bardgett and Wardle 2010). This is because aboveground and belowground communities are linked via their shared interactions with the plant community (Wardle et al. 2004; Wurst 2013). Consequently, ecosystem functions such as nutrient cycling will be correlated with aboveground and belowground community composition, which in turn will be correlated with plant community composition, including the expression of plant functional traits such as nutrient content and anti-herbivore defense (Bardgett and Wardle 2010; Schmitz et al. 2015; Burghardt 2016). Furthermore, aboveground-belowground food chain interactions and feedbacks on nutrient cycling may vary with environmental context, determined by variation in abiotic (e.g. climate, geology) and biotic (e.g. trophic cascades, community structure) conditions and variation in plant functional trait expression (Wardle et al. 2004; Eisenhauer and Scheu 2008; Andriuzzi and Wall 2017). However, observed correlations among system properties cannot be used to infer how specific biotic or abiotic drivers alter ecosystem functioning among contexts. Hence, making sense of this context-dependency requires manipulating individual variables that are expected to alter both aboveground-belowground functioning in the face of environmental change (Wardle and Bardgett 2004; Wardle et al. 2004; Dobson and Blossey 2015).
We report here on a landscape-scale study to evaluate how variation in aboveground and belowground communities is related to naturally occurring intraspecific variation in a functional trait—anti-herbivore chemical defence composition—of a shared plant species, Tansy (Tanacetum vulgare). The study is motivated by several lines of evidence that chemical defense ought to alter both aboveground and belowground communities. First, intraspecific variation in plant chemical defence is known to directly and/or indirectly influence the associated composition of arthropod communities (Wimp et al. 2005; Hughes et al. 2008; Schweitzer et al. 2008; Fischer et al. 2014). Second, variation in plant anti-herbivore defence traits are known to cause variation in plant nutrient quality across landscapes (Burghardt 2016). Third, plant anti-herbivore chemistry can reduce plant-litter decomposition rates (Schweitzer et al. 2008). Fourth, variation in plant defence can influence the composition of the soil microbial community (Bardgett and Wardle 2010). But, whether or not plant variants of a given chemical defense strategy have the same qualitative influence on both aboveground and belowground communities remains largely uncertain for most ecosystems (Bardgett and Wardle 2010; Schmitz et al. 2015) and especially so for the Tansey study system.
The purpose of this study was to resolve if and how variants in plant chemical defense, aboveground and belowground community structure and soil nutrient status are related. We used Tansy chemical defense composition as focus for our analysis, given that Tansy plants have different variants of dominant defense chemicals (e.g., thujon, borneal, camphor). We examined whether or not differences in chemical defense was a dominant mechanism that could modify aboveground and belowground community composition and nutrient cycling. Such an approach allows one to follow the influence of that focal variable with the potential to add key mechanistic understanding of the relationship (e.g. Buchkowski et al. 2018).
We focused on the aboveground arthropod communities and belowground microbial communities associated with Tansy, because these are expected to have the largest influence on the critical ecosystem process of herbivory and decomposition in our system (Holtkamp et al. 2011; Schmitz and Price 2011; Lehmann and Kleber 2015). We expected that the specific chemical defense composition of Tansy plants would be directly related to the composition of aboveground arthropod and belowground microbial communities, with less effective chemical defenses promoting higher arthropod and microbial diversity and use (Grime et al. 1996).
Material and methods
Study system
Tansy (Tanacetum vulgare) is a perennial herbaceous composite originating in Europe and Asia (Holopainen et al. 1987) that preferentially grows in disturbed, well-drained, poor soils (Keskitalo et al. 1998). It often forms isolated patches alongside river valleys, railway tracks and on abandoned lands. Single plants comprise a genet with up to 50 flowering shoots. Genets propagate clonally underground via stolons. Tansy contains a volatile oil composed of the chemicals β-thujon, camphor, and borneol that give the plant its bitter taste to vertebrates (Benedek et al. 2015; Bálint et al. 2016; Clancy et al. 2016). Different populations of Tansy, known as “chemotypes”, vary in the concentration of each of those chemicals. Indeed, breeding experiments and assays using molecular markers have demonstrated that the chemical oil produced by a particular Tansy plants are genetically determined (Holopainen et al. 1987; Keskitalo et al. 1998).
Three specialized aphid species feed on Tansy: Macrosiphoniella tanacetaria (Kaltenbach), Metopeurum fuscoviride Stroyan and Uroleuchon tanaceti L. M. tanacetaria (MA) a green aphid is not attended by ants and forms small colonies mainly on the top of shoots. M. fuscoviride (ME) is a black-purple aphid and obligatory ant-tended that feeds in more compact and often large colonies on leaves and near the apex of shoots. The black garden ant, Lasius niger (L.) commonly tends M. fuscoviride aphids and help to defend from the predators (Loxdale et al. 2011). The crimson tansy aphid U. tanaceti (UR), a red aposematic aphid that it is distasteful to predators, is found underneath leaves (Dransfield and Brightwell 2015). Many predators attack MA and ME aphids, the most important being the aphid specialist 7-spotted ladybird beetle (Coccinella septempunctata). Predation by the generalist nursery web spider (Pisaura mirabilis) and minute pirate bugs (Orius spp.) also occurs (Mehrparvar et al. 2013).
Selection of tansy plants for experiment
A previous study examined 100 tansy plants, sampled along a 120 km transect in Transylvania, Central Europe (Benedek et al. 2015; Bálint et al. 2016). These plants, relatively similar in size (~1.0–1.2 m high), were genotyped by RAPD procedures using 13 primers, following established methods (Keskitalo et al. 1998). The 100 plants represented by 30 genotypes. Each genotype grew as a unique population in different locations across the 150 km gradient, but they were not completely isolated from each other because all grew near a main road or railway. Together the 30 individual genotypes formed the basis for our field survey and soil analyses (Table 1 and Figure 1 and 2 supplementary online materials).
Field assessment of tansy aphids, its predators and mutualist ants
Our 2-year (2016 and 2017) field study was conducted at sites containing these 30 genotypes. There were no obvious differences in soil (well-drained, poor clay) or climate among the sites in which these plants were found. The climate was temperate, typical of the Sub-Carpathian region, with abundant precipitation in spring and fall. Summer relatively warm (18 to 30 °C) and cold (to −20 °C) in winter.
The distance between selected genotypes varied between 1 and 150 km. All plants were labelled individually and their spatial location was recorded using a GPS at the start of the assessment. Plants were examined for the duration of the study, which involved bi-weekly censuses between 7 and 11 a.m. from June when the aphids first appeared until the end of September when aphids had disappeared. All three aphid species and predatory arthropods found on marked plants were identified and counted. All MA and ME aphids were counted as individuals, except when the colony size of ME on a shoot was large. In this case, the spatial extent of the colony on the shoot was measured and all aphids within in a 5 cm section of the shoot were counted to estimate colony size. Because of their high density, and to minimize disturbances to plants, colonies of crimson aphids UR, which resided on the underside of leaves, were assessed every second week by photographing all colonies. High-resolution (20 megapixel Nikon Camera) images were obtained by turning over the leaves on which colonies were observed and taking photographs. UR aphid colonies were sampled on the same days as all other aphid and predator sampling. All photographs were saved, each identified by the plant’s identifying code. The exact number of aphids was counted in the lab from photos. After counting and photographing UR aphids in the field, a ten-minute visual scan was made of all other aphid colonies on all plants. Furthermore, counts were made of all ant workers and all aphid predators observed attending and within 1 cm of the aphid colony.
Volatile extraction from tansy plants and quantitative analysis of volatiles
In early June of the first year, 100 g of live plant material (fresh leaves and stems) was collected from each of the 30 marked tansy plants. Samples were collected from ramets unoccupied by aphids. These samples were used to analyse the chemical composition of each individual plant using GC-MS (Table 1 supplementary online materials). GC-MS analysis was performed using a chromatograph (Agilent 7890B GC interfaced to Agilent 5977) and a mass spectrometer (ionization voltage 70 eV) equipped with a HP-5MS 30 × 0.25 mm capillary column. Volatiles were extracted by holding at 50 °C for 2 min, then systematically increasing from 60 °C - 200 °C at a rate of 5 °C min − 1, holding for 1 min, then systematically increasing temperature further from 200 to 240 °C at a rate of 10 °C min − 1, using He as the carrier gas (1.0 mL min − 1). The temperatures of the injector and detector were 220 and 290 °C, respectively. Qualitative analysis was based on a comparison of retention times and indexes on both columns and mass spectra with corresponding data in computer mass spectra libraries (Nist14). All 30 tansy genotypes were then identified to chemotype based on their volatile terpene-terpenoid profile and categorized as ‘pure’ chemotypes when a single chemical made up 90% or more of the total quantity of compounds detected, e.g. “β-Thujon pure”. ‘Hybrid’ chemotypes were those in which the dominant volatile made up more than 40% of the composition, but the next most frequent was at least 20% lower in quantity than the dominant one. In this case, the plant was named as “β-Thujon hybrid”. When two compounds accounted for >60% of the plant volatiles and differences were less than 10% between volatiles, plants were designated by the dominant compound present e.g. “β-Thujon mixed”. When three compounds accounted together >75% of the plant volatiles (one plant) or when we were unable to determine the whole volatile composition of the plant, these were named as ‘unclassified’ chemotypes (one plant) (Table 1 supplementary online materials) see also (Benedek et al. 2015; Bálint et al. 2016).
Soil analyses and litter decomposition experiment from soil of different tansy chemotypes
Soil analyses were made after the first year of arthropod assessments at each of the 30 Tansy sites by collecting 100 g at both 5 cm and 10 cm below each of the 30 plants. The soil type was Stagnic Argic Luvisol formed on Holocene loamy-clay alluvial-proluvial deposits developed on a quasi-horizontal surface covered by spontaneous flora elements. The upper soil horizon had 3.26% humus, mobile potassium content of 408 ppm, and total nitrogen varying between 0.143% and 0.101%. The base saturation of the upper layer was 77.85%, and the pH (H2O) 6.38, decreasing with soil depth. At depth (35–73 cm) the soil layer was comprised of mixed clay accumulation carrying high amounts of available nutrient ions.
Upon completing soil analyses, site locations supporting three similar sized naturally growing Tansy plants of the same chemotype (i.e., three individuals dominated by Camphor, three by Borneol and three by β-Thujon) were selected for further analyses. Replicate plants were identified along a gradient of 2000 m, separated by at least 500 m, ensuring that the replicate plants occupied sites with the same total soil N contents. For the duration of the study, all plant litterfall not belonging to the focal Tansy plant was removed from a three metre area surrounding the plant. This ensured that the soil would receive only Tansy litterfalls. In November soil was collected to a 10 cm depth below each tansy chemotype. Soils were sterilized in the lab. Then, collected Tansy litterfall from each chemotype was mixed with soil in the proportion 1/3 litterfall and 2/3 soil. The mixture was then placed in litterbags for decomposition analysis. Because the litter bag mesh size may hamper litter decomposition due to interference with agents of litter fragmentation (Lecerf 2017), we used bags with 0.2 mm mesh. At the end of November 2016, four litter bags each containing 100 mg of soil-litter mixture were placed under each Tansy chemotype at a depth of 10 cm in the soil, to enable inoculation by the resident, natural microbial community. Beginning at the end of February 2017 and repeating at the end of March, April and May, we collected one litterbag from under each chemotype. Litterbag samples were placed in sterile tubes and stored at −70 °C until DNA analyses. To control for effects of just Tansy litter, we additionally created litterbags with mixtures of soil and natural vegetation (dominated by monocots) from plants other than Tansy. Four bags were placed underneath each Tansy chemotype, alongside the Tansy litterbags.
DNA sequencing of microbial community from soil of different tansy chemotypes
Environmental genomic DNA was extracted from all soil samples using a DNeasy PowerSoil Kit (Qiagen) following manufacturer’s instructions, with the exception that the cell disruption step involved shaking each sample at 30 Hz for 3 min using a Mixer Mill MM301 (Retsch). A part of the 16S rRNA gene was amplified with fusion primers containing the Bacteria-specific sequences Bakt_341F (5’-CCT ACG GGN GGC WGC AG-3′) and modified Bakt_805R (5’-GAC TAC NVG GGT ATC TAA TCC-3′) (Herlemann et al. 2011), respectively, and Fluidigm CS1 and CS2 universal sequences at their 5′ ends. In addition samples were sent to the Genomics Core, Research Technology Support Facility, Michigan State University (USA) for further analysis. There, illumina compatible, dual indexed adapters were added by PCR with primers targeting the CS1 and CS2 sites. PCR products were then batch normalized using SequalPrep DNA Normalization plates (Invitrogen) and all product recovered from the normalization plate was pooled. Subsequently a clean-up of this pool was performed with Agencourt AMPure XP magnetic beads (Beckman Coulter). Quality control and quantification was carried out using a combination of Qubit dsDNA HS (Thermo Fisher Scientific), Fragment Analyzer High Sensitivity DNA (Advanced Analytical) and Kapa Illumina Library Quantification qPCR (Kapa Biosystems) assays. The pool was then loaded onto a standard MiSeq v2 flow cell (Illumina). Sequencing was performed in a 2 × 250 bp paired end format using a v2, 500 cycle MiSeq reagent cartridge. Custom sequencing and index primers complementary to the CS1/CS2 oligomers were added to appropriate wells of the reagent cartridge. Base calling was done by Illumina Real Time Analysis (RTA) v1.18.54 and output of RTA was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v2.19.1.
Tansy chemotypes and soil total N analyses
Nitrogen availability to aphids was measured in each of the Tansy chemotypes used for litterbag decomposition analyses. Plant material (100 g of fresh leaves and shoots unoccupied by aphids) was first collected in May, after litterbags were removed, and then whenever aphids and predators were sampled. Soil samples from 10 cm deep were collected at the same time when arthropods were sampled. N content of soil from litterbags used for bacterial community assessment were also analysed. N content for plants was determined using an HCN elemental-analyser (Bird et al. 2017). Soil total available N and litterbags soil N were measured using Kjeldahl approaches (Kjeldahl 1883; Wang et al. 2015).
Data analysis - field data
Arthropod data analyses began by characterising the species composition and abundance for each individual chemotype. We tested for inter-annual differences in abundance of each arthropod species by constructing principal response curves (Van den Brink and Braak 1999), which account for population dynamics through the season. Mean number of arthropods per individual plant per sampling date was estimated. Analyses revealed that only pure β-Thujon and one hybrid of Carvone had very low numbers or no arthropods. Likewise, two ‘unclassified’ chemotypes also had few or no arthropods. Therefore these four plants were not considered in further analyses. That left 26 mixed chemotypes available for further arthropod analyses.
Aphids, ants, and predators were consistently present together on the mixed chemotypes from June 4 until September 12 of the first year, and from June 5 until September 14 of the second year. Hence, the nine bi-weekly samples collected were used for analysis. We tested for a year effect by comparing the mean number of arthropods on each plant using a repeated measures analysis on bi-weekly sampling dates between years. No significant temporal difference, either within or between years, was detected. Therefore we pooled data for both years in all subsequent analyses.
We tested for the effect of plant chemotype profile (considering all volatiles according to their relative proportion in a Tansy plant) on different attributes of arthropod abundances. This was done by first subjecting data on the relative amount of each chemical per plant (%) to a Principal Components Analyses (PCA) to identify the proportion of variation in each PCA axis and was explained also by each dominant chemical (Camphor, Borneol and β-Thujon). PCA covariance analyses were run using Community Analysis Package 4 in R(R Core Team 2013). The average count of each invertebrate grouping was used as a response variable in regressions with PCA axis1 (PCA1), PCA axis 2 (PCA2) and PCA axis 3 (PCA3) scores as predictor variables with F and p values computed at 99.5% confidence intervals.
Bioinformatic and statistical analyses of microbial community
Bioinformatic analysis of the resulting soil sample sequence reads were carried out with mothur v1.38.1 (Schloss et al. 2009) using the MiSeq SOP (http://www.mothur.org/wiki/MiSeq_SOP downloaded at 04/04/2018) (Kozich et al. 2013) with the following exceptions: ‘deltaq’ adjusted to 10 in the ‘make.contigs’ command, UCHIME (Edgar et al. 2011) was used for chimera detection, and singleton reads were also removed from the dataset according to (Kunin et al. 2010). Taxonomic assignments were made based on the ARB-SILVA SSU Ref NR 132 (Quast et al. 2013), with a minimum bootstrap confidence score of 80%, and using 1000 iterations.
Microbial species richness was estimated by applying a 0.15 cutoff in mothur’s ‘dist.seqs’ command and subsequently using 97% 16S rRNA gene sequence similarity, corresponding to the prokaryotic species-level threshold recommended by (Tindall et al. 2010). Microbial alpha diversity (estimated using the Shannon-Wiener and Inverse Simpsons’s (1/D) diversity indices) and species richness values (using the Chao1 and the ACE richness metrics) were calculated using mother v1.38.1. Linear regression was used to assess the variation in total bacterial diversity indices (Shannon and Simpson) under Camphor, Borneol and β-Thujon chemotypes and control in four litterbag sampling periods (February, March, April and May), and R2 values computed using PAST (Figures 3, 4 and 5 Supplementary online materials). Variation in bacterial community composition was also compared between genera for each tansy chemotype, control and mineralization process (from February to May) with ANOVA using mean percentages of DNA from total samples.
Seasonal microbial community composition was related to variations in plant chemical content using factorial analyses with microbial DNA diversity values and related diversity in plant defense chemistry transformed to a log10 scale.
The plant and soil available N content was compared between Tansy chemotypes (Camphor, Borneol and β-Thujon) using ANOVA and Tukey test at 99.5% confidence intervals.
Results
Tansy genotypes distribution by chemical compounds
Of the 30 Tansy genotypes examined for chemical composition one was a pure chemotype, with 99% of β-Thujon. Another 19 genotypes were hybrid chemotypes, including: one Carvone hybrid, 12 β-Thujon hybrids, three Borneol hybrids and three Camphor hybrid plants. A further eight plants were mixed chemotypes, with β-Thujon, Camphor and Borneol being the most dominant compounds identified in those genotypes. Two plants were unclassified because more than two chemicals, each between 20 and 30%, represented the chemotypes (one plant), or we were unable to detect dominant chemicals (one plant). Altogether, Thujon plants dominated the site (Table 1, Figure 1 supplementary online materials). PCA confirmed that the greatest variation in chemical profile among individual plants was explained primarily by the relative amount of Thujon in the plants (PCA1 and PCA2) followed by Camphor and Borneol (PCA1 and PCA2). Also, a low effect of another chemical Carvone was detected (Fig. 1, Table 2 Supplementary online materials).
PCA of tansy chamotypes using the variation in chemical profile among individual plants. The dominant profiles were explained by the relative amount of Thujon, Camphor and Borneol. Green ellipse represents Thujon plants, blue ellipse represents Camphor plants, red ellipse represents Borneol plants and gold ellipse represents Carvone plants while grey ellipse other plants with no clear chemotype profiles
Arthropod density variation by tansy chemical compounds
β-Thujon and Camphor together had a significant negative effect on UR aphids and spiders and a positive effect on seven-spotted ladybird (Table 1). The abundance of ME and MA aphids and spiders was to some extent positively influenced by Borneol and β-Thujon together (PCA2). We detected a significant effect of the dominant compounds on most arthropod abundances, except ants (Table 1). Significantly more ME, MA, UR and ladybird beetles, but lower numbers of spiders, were detected on Camphor dominated plants. Borneol had positive effects on ME, UR aphids and spiders. There was a significant negative effect of β-Thujon on the number of all arthropods, except ants where no effect was detected (Table 1). Three predator species were often observed on plants: the seven-spotted ladybird (C. septempunctata), the nursery web spider (P. mirabilis) and several species of minute pirate bugs (Orius spp.). Seven-spotted ladybird beetles were more abundant on Camphor chemotypes and their abundance was positively influenced by ME aphids and negatively influenced by spiders. Spider abundance was positively associated with both ME and MA aphids, and Orius, and negatively associated with ladybird beetles. The abundance of Orius was positively associated with ME. Ants always tended Tansy chemotypes with high ME colonies (Table 1).
Soil bacterial community variation by tansy chemical compounds
The Illumina Real Time analysis and soil bacterial taxonomic assignments (ARB-SILVA SSU Ref NR 132) revealed that the total estimated soil microorganism species varied between Tansy chemotypes. Bacterial diversity indices decreased in litterbags under β-Thujon chemotypes (Table 2). Considering the most frequent bacterial species, Flavobacterium dominated the microbial community decomposing litter under Camphor chemotypes during March. Flavobacterium also dominated litter decomposing microbes under β-Thujon chemotypes in April and May (Figs. 2 and 3a, b, c). Dominance of Chloroflexi and Gaiella in control were observed except April when also high amount of Flavobacterium DNA were detected (Fig. 3d). Linear regression also revealed that the frequency of most important bacterial genera in litter biodegradation (Flavobacterium, Pedobacter, Pseudomonas, Massilia, Sphingomonas) generally decreased with time in soil of Camphor and Borneol, while there was a significant increase of Flavobacterium and decrease of Sphingomonas and Pseudomonas in soil of β-Thujon (Figure 3, 4 and 5 supplementary online materials). This was also different in control, where significant increase of Pedobacter, Chloroflexi, Sphingomonas, Pseudomonas and Acidobacteria were detected (Figure 4 supplementary online materials). Generally lower species numbers were estimated in soils associated with Thujon chemotypes, even lower than in controls (Fig. 3, Table 2).
Bacterial diversity indices in litterbags (a), estimated after next-generation DNA sequencing, MiSeq v2 flow cell (Illumina) and the frequency of the dominant bacteria (b) in soil associated with different Tansy chemotypes and the non-Tansy control over the course of the study (from February to May). Taxonomic assignments were based on the ARB-SILVA SSU Ref NR 132 with a minimum bootstrap confidence score of 80%, and using 1000 iterations. Acidobacteria, Chloroflexi Pedosphaeraceae and Saccharimonadales were not identified to genus level
Variation in the dominant bacteria in soil associated with different Tansy chemotypes and the non-Tansy control over the course of the study (from February to May). Taxonomic assignments were based on the ARB-SILVA SSU Ref NR 132 with a minimum bootstrap confidence score of 80%, and using 1000 iterations. Acidobacteria, Chloroflexi Pedosphaeraceae and Saccharimonadales were not identified to genus level
A clear seasonal turnover in microbial community structure was detected. A dominance of Sphingomonas, Massilia, Pseudomonas and Pedobacter was detected in soil of all chemotype in February and March, while Sacharimonadales, Gaielle, Chloroflexi, Acidobacteria and Pedosphaeraceae dominated each Tansy chemotype soils in April and May. These species were also dominant in control soil without Tansy from February to May (Fig. 4).
Shannon H and Inverse Simpson diversity of the total bacterial community increased with amount of litter decomposition in soils associated with Camphor and Borneol chemotypes (Figure 4 supplementary online materials). This was not observed in soil of β-Thujon. Also higher total bacterial community diversity was detected in the control that did not contain Tansy litterfall (Figure 5 supplementary online materials).
Tansy plant N and soil N content variations
Significant differences in N content of soils below chemotypes N (FCamphor-Borneol = 5.6, P < 0.001; FCamphor-Thujon = 10.9, P < 0.001; FThujon-Borneol = 5.4, P < 0.001) and also differences in plant N content (FCamphor-Borneol = 4.9, P < 0.001; FCamphor-Thujon = 9.3, P < 0.001; FThujon-Borneol = 4.3, P < 0.001) were detected (Fig. 5a, b). Litterbag N content was significantly higher in litterbags under Camphor, followed by Borneol and least in litterbags under Thujone and control (FCamphor-Borneol = 4.3, P < 0.001; FCamphor-Thujon = 9.1, P < 0.001; FThujon-Borneol = 4.1, P < 0.001, FCamphor-Control = 9.2, P < 0.001; FBorneol-Control = 3.3, P < 0.01; FThujon-Control = 0.9, P < 0.11) (Fig. 5c).
Discussions
The chemotype profile of Tansy genotypes shows that β-Thujon dominated the study area (17 plants out of 30 of which 13 plants had high dominance of β-Thujon, and four other plants with high β-Thujon content) (Table 1 and Figure 1 supplementary online materials). The only pure chemotype with 99% β-Thujon was the furthest away from Borneol and Camphor plants, and it was surrounded only by other β-Thujon dominated plants. This same pattern was revealed in another study carried out on 100 tansy plants (Bálint et al. 2016). This suggest that β-Thujon may have evolutionary importance as antifeedant compound and may protect Tansy plants against herbivores. This conclusion can also be inferred from patterns of variation in arthropod densities among chemotypes.
The abundance of all aphids and predators was negatively associated with β-Thujon dominance, while a generally positive association of all arthropods except ants was observed with Borneol and Camphor dominated Tansy plants (Table 1). Previous studies on thyme demonstrated a similar role of plant chemical content. The negative effects of spider and ladybird predators on aphids in turn varied by plant chemical content (Linhart et al. 2005). Aphid (Aphis serpylli) density on thyme plants with Carvacrol, Geraniol and Thymol monoterpes were reduced by spider and ladybird predators. Aphid colonies on Linalol dominated plants however were not affected by these predators, probably because predators showed chemotype-specific feeding behaviour and avoided Linalol plants (Linhart et al. 2005).
The soil bacterial community composition also varied highly between Tansy chemotypes. For Camphor and Borneol chemotypes the diversity of the total bacterial community increased significantly during the temporal sequence of litterbag sampling. This compositional shift was not observed in the soil from β-Thujon plants. The frequency of bacterial genera with a significant role in litter biodegradation and mineralization processes (Flavobacterium, Pedobacter, Pseudomonas, Massilia, Sphingomonas) clearly decreased with time in soil of Camphor and Borneol. This did not happen in soil of β-Thujon where the dominance of Flavobacterium and decrease of Sphingomonas and Pseudomonas was significant during the mineralization process (Figures 2, 3 and Fig 4 supplementary online materials). Pedobacter, Pseudomonas, Massilia, Sphingomonas are the most important genera for litter degradation and mineralization. Their role in early decomposition process has been demonstrated (Purahong et al. 2016). The early dominance of these microorganisms in soil of Camphor and Borneol plants is indicative of fast litter decomposition and N mineralization, leading to higher mineral and macronutrient (N in this case) content available for these plants. Flavobacterium (phylum Bacteriodetes) species participate in the aerobic biodegradation of macromolecules such as terpenoids. Hence their high dominance on soil of β-Thujon plants can be explained by fact that β-Thujon is toxic to other organisms, including mammals but also microorganisms (Lachenmeier and Uebelacker 2010). Therefore, decomposition and N mineralization may be lower over time. High variation of Pedobacter, Pseudomonas and Sphingomonas frequency was detected between chemotypes.
During the early mineralization period in soil of β-Thujon these genera reach 15–20% compared with 5–10% in soil beneath Camphor and Borneol. But in April and May, the dominance of Flavobacterium in β-Thujon suggest a much lower decomposition and mineralization process, hence a lower nutrient content of β-Thujon dominated plants (Figs. 3 and 4). Massilia may also have importance in fast mineralization process the variation in frequency during the mineralization process were significant on soil of Borneol and Camphor and less obvious in soil of β-Thujon. Massilia are aerobic chemoorganotrophs (Baldani et al. 2014). Members of the genus promote plant growth, and are important members of the rhizosphere (Ofek et al. 2012), while their significance during the decay of plant biomass was also reported (Moura et al. 2018). The variation of the other dominant microorganisms (Chloroflexi, Gaiella and Acidobacteria) were less obvious, but their role in Tansy litter decomposition and mineralization cannot be discounted. For example the genus Gaiella (phylum Actinobacteria) is strictly aerobic, growing at neutral pH and is capable of utilizing various sugars and amino acids (Albuquerque et al. 2011). These bacteria are abundant in different soils even in those included those associated with plants (Correa-Galeote et al. 2016).
Altogether our results show that Tansy litter decomposition and N mineralization follow different pathways and is highly influenced by the plant dominant chemical compounds. Because N mineralization follows a faster process in soil of Camphor and Borneol plants, the availability of the minerals and macro elements (N) from soil should be high. A much lower process rate in soil of Thujon chemotypes occurs, which should result in lower amounts of nutrient available to these plants. This is corroborated by soil and plant N content analyses, where Thujon chemotypes had significantly lower available soil N and plant tissue N content than the other chemotypes (Fig. 5). Altogether Tansy chemistry has a decided impact on decomposition and nutrient recycling. Because Tansy is a perennial, these chemical effects on belowground decomposition process may have lasting effects on above-ground arthropod density.
In particular, predator associations were specific to different Tansy chemotypes. The aphid specialist ladybird-beetles were significantly and predominantly associated with Camphor while significantly higher numbers of the polyphagous nursery web spider was observed on Borneol plants. There was no specific association of these predators with Thujon chemotypes. The basis for these associations has been partially discussed previously (Bálint et al. 2016). We identify here additional mechanisms.
While earlier work shows that differences in ME aphid abundance and attraction of predators can be attributed to different specific volatile (Benedek et al. 2015; Bálint et al. 2016), we show additionally that high variation in N tissue content between Tansy chemotypes seems to have important drivers of host quality because of the potential nutritional benefits to some prey species (ME and MA aphids in this case). This variation in bottom-up effect in turn influences predator density on the different Tansy chemotypes. Altogether we detected that the intra-specific chemical variation of a single plant species has significant influence in litter decomposition and N mineralization processes and, through nutrient allocation, influences above-ground arthropod food-web structure. The present study also demonstrates a range community-level outcomes between soil microorganisms-plant-arthropod networks that are highly influenced by plant chemical content and has important eco-evolutionary effects through lower phytophagous insect density and higher dominance of some chemotypes. These mechanisms can partially explain the dominance and spatial distribution of Tansy plants with Thujon dominance as compound.
In sum, we have resolved the interplay between variation in the nature and level of plant defence expression within genotypes of a plant species regulated by available soil nutrients to plants, which is in turn determined by soil microbial community composition and the degree and rate to which microbes can decompose and release nutrients from detrital inputs from defended plants. Resolving this chain of effect enhances the ability to predict spatial variation in plant population structure and associated arthropod species structure and ecosystem functioning.
References
Albuquerque L, Rainey FA, Nobre MF, da Costa MS (2011) Schleiferia thermophila gen. nov., sp. nov., a slightly thermophilic bacterium of the phylum “Bacteroidetes” and the proposal of Schleiferiaceae fam. nov. Int J Syst Evol Microbiol 61:2450–2455. https://doi.org/10.1099/ijs.0.028852-0
Andriuzzi WS, Wall DH (2017) Responses of belowground communities to large aboveground herbivores: meta-analysis reveals biome-dependent patterns and critical research gaps. Glob Chang Biol 23:3857–3868
Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD (2014) The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384:413–431. https://doi.org/10.1007/s11104-014-2186-6
Bálint J, Zytynska SE, Salamon RV, Mehrparvar M, Weisser WW, Schmitz OJ, Benedek K, Balog A (2016) Intraspecific differences in plant chemotype determine the structure of arthropod food webs. Oecologia 180:797–807. https://doi.org/10.1007/s00442-015-3508-y
Bardgett RD, Wardle DA (2010) Aboveground-belowground linkages: biotic interactions, ecosystem processes, and global change. Oxford University Press, New York
Benedek K, Bálint J, Salamon RV, Kovács E, Ábrahám B, Fazakas C, Loxdale HD, Balog A (2015) Chemotype of tansy (Tanacetum vulgare L.) determines aphid genotype and its associated predator system. Biol J Linn Soc Lond 114:709–719. https://doi.org/10.1111/bij.12445
Bird M, Keitel C, Meredith W (2017) Analysis of biochars for C, H, N, O and S by elemental analyser. In: Singh B, Camps-Arbestain M, Lehmann J (eds) Biochar: a guide to analytical methods. CSIRO Publishing, Melbourne, VIC, Australia, pp 39–50
Buchkowski RW, Schmitz OJ, Bradford MA (2018) Nitrogen recycling in coupled green and brown food webs: Weak effects of herbivory and detritivory when nitrogen passes through soil. J Ecol 107:963–976. https://doi.org/10.1111/1365-2745.13079
Burghardt KT (2016) Nutrient supply alters goldenrod’s induced response to herbivory. Funct Ecol 30:1769–1778. https://doi.org/10.1111/1365-2435.12681
Clancy MV, Zytynska SE, Senft M, Weisser WW, Schnitzler JP (2016) Chemotypic variation in terpenes emitted from storage pools influences early aphid colonisation on tansy. Sci Rep 6:38087. https://doi.org/10.1038/srep38087
Correa-Galeote D, Bedmar EJ, Fernández-González AJ, et al (2016) Bacterial communities in the rhizosphere of Amilaceous maize (Zea mays L.) as assessed by pyrosequencing. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.01016
Dobson A, Blossey B (2015) Earthworm invasion, white-tailed deer and seedling establishment in deciduous forests of North-Eastern North America. J Ecol 103:153–164. https://doi.org/10.1111/1365-2745.12350
Dransfield RD, Brightwell R (2015) Colour in aphids - aposematic, cryptic or both? Antenna (Bulletin of the Royal Entomological Society) 39(2):60–71
Edgar EC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Eisenhauer N, Scheu S (2008) Earthworms as drivers of the competition between grasses and legumes. Soil Biol Biochem 40:2650–2659
Fischer DG, Chapman SK, Classen AT, Gehring CA, Grady KC, Schweitzer JA, Whitham TG (2014) Plant genetic effects on soils under climate change. Plant Soil 379:1–19. https://doi.org/10.1007/s11104-013-1972-x
Grime JP, Cornelissen J, (Hans) HC, Thompson K, Hodgson JG (1996) Evidence of a causal connection between anti-herbivore Defence and the decomposition rate of leaves. Oikos 77:489. https://doi.org/10.2307/3545938
Herlemann DP, Labrenz M, Jürgens K et al (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. https://doi.org/10.1038/ismej.2011.41
Holopainen M, Hiltunen R, Lokki J et al (1987) Model for the genetic control of thujone, sabinene and umbellulone in tansy (Tanacetum vulgare L.). Hereditas 106:205–208. https://doi.org/10.1111/j.1601-5223.1987.tb00254.x
Holtkamp R, van der Wal A, Kardol P, van der Putten WH, de Ruiter PC, Dekker SC (2011) Modelling C and N mineralisation in soil food webs during secondary succession on ex-arable land. Soil Biol Biochem 43:251–260. https://doi.org/10.1016/j.soilbio.2010.10.004
Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M (2008) Ecological consequences of genetic diversity. Ecol Lett 11:609–623. https://doi.org/10.1111/j.1461-0248.2008.01179.x
Keskitalo M, Lindén A, Valkonen JPT (1998) Genetic and morphological diversity of Finnish tansy (Tanacetum vulgare L., Asteraceae). Theor Appl Genet 96:1141–1150. https://doi.org/10.1007/s001220050850
Kjeldahl J (1883) Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern. Fresenius, Zeitschrift f anal Chemie 22:366–382. https://doi.org/10.1007/BF01338151
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. https://doi.org/10.1128/AEM.01043-13
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P (2010) Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12:118–123. https://doi.org/10.1111/j.1462-2920.2009.02051.x
Lachenmeier DW, Uebelacker M (2010) Risk assessment of thujone in foods and medicines containing sage and wormwood--evidence for a need of regulatory changes? Regul Toxicol Pharmacol 58:437–443. https://doi.org/10.1016/j.yrtph.2010.08.012
Lecerf A (2017) Methods for estimating the effect of litterbag mesh size on decomposition. Ecol Model 362:65–68. https://doi.org/10.1016/j.ecolmodel.2017.08.011
Lehmann J, Kleber M (2015) The contentious nature of soil organic matter. Nature 528:60–68. https://doi.org/10.1038/nature16069
Linhart YB, Keefover-Ring K, Mooney KA, Breland B, Thompson JD (2005) A chemical polymorphism in a multitrophic setting: thyme monoterpene composition and food web structure. Am Nat 166:517–529. https://doi.org/10.1086/444438
Loxdale HD, Schöfl G, Wiesner KR et al (2011) Stay at home aphids: comparative spatial and seasonal metapopulation structure and dynamics of two specialist tansy aphid species studied using microsatellite markers. Biol J Linn Soc 104:838–865. https://doi.org/10.1111/j.1095-8312.2011.01761.x
Mehrparvar M, Mahdavi Arab N, Weisser WW (2013) Diet-mediated effects of specialized tansy aphids on survival and development of their predators: is there any benefit of dietary mixing? Biol Control 65:142–146. https://doi.org/10.1016/j.biocontrol.2012.12.009
Moore JC, Berlow EL, Coleman DC, Ruiter PC, Dong Q, Hastings A, Johnson NC, McCann KS, Melville K, Morin PJ, Nadelhoffer K, Rosemond AD, Post DM, Sabo JL, Scow KM, Vanni MJ, Wall DH (2004) Detritus, trophic dynamics and biodiversity. Ecol Lett 7:584–600. https://doi.org/10.1111/j.1461-0248.2004.00606.x
Moura BB, Hoshika Y, Silveira NM, Marcos FCC, Machado EC, Paoletti E, Ribeiro RV (2018) Physiological and biochemical responses of two sugarcane genotypes growing under free-air ozone exposure. Environ Exp Bot 153:72–79. https://doi.org/10.1016/j.envexpbot.2018.05.004
Ofek M, Hadar Y, Minz D (2012) Ecology of root colonizing Massilia (Oxalobacteraceae). PLoS One 7:e40117. https://doi.org/10.1371/journal.pone.0040117
Purahong W, Wubet T, Lentendu G, Schloter M, Pecyna MJ, Kapturska D, Hofrichter M, Krüger D, Buscot F (2016) Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol Ecol 25:4059–4074. https://doi.org/10.1111/mec.13739
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/nar/gks1219
R Core Team (2013) R: A language and environment for statistical computing. Vienna, Austria. The R Foundation for Statistical Computing. ISBN: 3-900051-07-0. Available online at http://www.R-project.org/. Accessed 2 Aug 2019
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Schmitz OJ, Price JR (2011) Convergence of trophic interaction strengths in grassland food webs through metabolic scaling of herbivore biomass. J Anim Ecol 80:1330–1336. https://doi.org/10.1111/j.1365-2656.2011.01882.x
Schmitz OJ, Buchkowski RW, Burghardt KT, Donihue CM (2015) Functional traits and trait-mediated interactions: connecting community-level interactions with ecosystem functioning. Adv Ecol Res 319–343
Schweitzer JA, Madritch MD, Bailey JK, LeRoy CJ, Fischer DG, Rehill BJ, Lindroth RL, Hagerman AE, Wooley SC, Hart SC, Whitham TG (2008) From genes to ecosystems: the genetic basis of condensed tannins and their role in nutrient regulation in a Populus model system. Ecosystems 11:1005–1020. https://doi.org/10.1007/s10021-008-9173-9
Tindall BJ, Rosselló-Móra R, Busse H-J et al (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266. https://doi.org/10.1099/ijs.0.016949-0
Van den Brink PJ, Braak CJFT (1999) Principal response curves: analysis of time-dependent multivariate responses of biological community to stress. Environ Toxicol Chem 18:138–148. https://doi.org/10.1002/etc.5620180207
Wang D, Chakraborty S, Weindorf DC, Li B, Sharma A, Paul S, Ali MN (2015) Synthesized use of VisNIR DRS and PXRF for soil characterization: Total carbon and total nitrogen. Geoderma 243–244:157–167. https://doi.org/10.1016/j.geoderma.2014.12.011
Wardle DA, Bardgett RD (2004) Human-induced changes in large herbivorous mammal density: the consequences for decomposers. Front Ecol Environ 2:145–153. https://doi.org/10.1890/1540-9295(2004)002[0145:HCILHM]2.0.CO;2
Wardle DA, Bardgett RD, Klironomos JN et al (2004) Ecological linkages between above ground and below ground biota. Science 304:1629–1633. https://doi.org/10.1126/science.1094875
Wimp GM, Martinsen GD, Floate KD, Bangert RK, Whitham TG (2005) Plant genetic determinants of arthropod community structure and diversity. Evolution 59:61–69
Wurst S (2013) Plant-mediated links between detritivores and aboveground herbivores. Front Plant Sci 4:10.3389
Acknowledgements
This research was funded by the Institute of Research Programs of the Sapientia Hungarian University of Transylvania, grant number 1/13/05.01.2012. The Illumina Real Time Analyser and soil bacterial taxonomic assignments (ARB-SILVA SSU Ref NR 132) studies were founded by PADOK 2/2017.
Funding
Open access funding provided by Sapientia Hungarian University of Transylvania (EMTE).
Author information
Authors and Affiliations
Contributions
AB and KB conceived the experiments. AB, KB, IM, CsF and JB designed and performed the experiments. GyM and CsA made the chemical analyses of tansy plants. CsF and IM made the soil analyses. IM, TF and ASz made the genetic assessment and analyses of soil samples. OJS, RWB, and AB analysed the data. AB, RWB and OJS wrote the manuscript. RWB and OJS provided editorial advice.
Corresponding authors
Ethics declarations
Conflict of interests
Authors declare no conflict of interests.
Additional information
Responsible Editor: Jeff R. Powell.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1039 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Benedek, K., Bálint, J., Máthé, I. et al. Linking intraspecific variation in plant chemical defence with arthropod and soil bacterial community structure and N allocation. Plant Soil 444, 383–397 (2019). https://doi.org/10.1007/s11104-019-04284-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04284-7