Abstract
In barley and other higher plants, phosphate homeostasis is maintained by a regulatory network involving the PHO2 (PHOSPHATE2) encoding ubiquitin-conjugating (UBC) E2 enzyme, the PHR1 (PHOSPHATE STARVATION RESPONSE 1) transcription factor (TF), IPS1 (INDUCED BYPHOSPHATESTARVATION1) RNA, and miR399. During phosphate ion (Pi) deprivation, PHR1 positively regulates MIR399 expression, after transcription and processing mature miR399 guides the Ago protein to the 5′-UTR of PHO2 transcripts. Non-coding IPS1 RNA is highly expressed during Pi starvation, and the sequestration of miR399 molecules protects PHO2 mRNA from complete degradation. Here, we reveal new cis- and trans-regulatory elements that are crucial for efficient PHO2 gene expression in barley. We found that the 5′-UTR of PHO2 contains two PHR1 binding sites (P1BSs) and one Pi-responsive PHO element. Using a yeast one-hybrid (Y1H) assay, we identified two candidate proteins that might mediate this transcriptional regulation: a barley PHR1 ortholog and a TF containing an uncharacterized MYB domain. Additional results classified this new potential TF as belonging to the APL (ALTERED PHLOEM DEVELOPMENT) protein family, and we observed its nuclear localization in barley protoplasts. Pi starvation induced the accumulation of barley APL transcripts in both the shoots and roots. Interestingly, the deletion of the P1BS motif from the first intron of the barley 5′-UTR led to a significant increase in the transcription of a downstream β-glucuronidase (GUS) reporter gene in tobacco leaves. Our work extends the current knowledge about putative cis- and trans-regulatory elements that may affect the expression of the barley PHO2 gene.
Key Message
The 5′-UTR of the barley PHOSPHATE 2 gene contains two P1BS motifs that can bind the transcription factor (TF) PHR1 (PHOSPHATE STARVATION RESPONSE 1) and the newly identified TF APL (ALTERED PHLOEM DEVELOPMENT)




Similar content being viewed by others

References
Abel S, Ticconi CA, Delatorre CA (2002) Phosphate sensing in higher plants. Physiol Plant 115(1):1–8. https://doi.org/10.1034/j.1399-3054.2002.1150101.x
Alaba S, Piszczalka P, Pietrykowska H, Pacak AM, Sierocka I, Nuc PW, Singh K, Plewka P, Sulkowska A, Jarmolowski A, Karlowski WM, Szweykowska-Kulinska Z (2015) The liverwort Pellia endiviifolia shares microtranscriptomic traits that are common to green algae and land plants. New Phytol 206(1):352–367. https://doi.org/10.1111/nph.13220
Aleksza D, Horváth GV, Sándor G, Szabados L (2017) Proline accumulation is regulated by transcription factors associated with phosphate starvation. Plant Physiol 175:555–567. https://doi.org/10.1104/pp.17.00791
Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LO (2012) Before it gets started: regulating translation at the 5′ UTR. Comp Funct Genomics 2012:475731. https://doi.org/10.1155/2012/475731
Bari R, Datt Pant B, Stitt M, Scheible WR (2006) PHO2, MicroRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999. https://doi.org/10.1104/pp.106.079707
Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6:673–685. https://doi.org/10.1046/j.1365-313X.1994.6050673.x
Bianchi M, Crinelli R, Giacomini E, Carloni E, Magnani M (2009) A potent enhancer element in the 5′-UTR intron is crucial for transcriptional regulation of the human ubiquitin C gene. Gene 448:88–101. https://doi.org/10.1016/j.gene.2009.08.013
Bonke M, Thitamadee S, Mahonen AP, Hauser MT, Helariutta Y (2003) APL regulates vascular tissue identity in Arabidopsis. Nature 426:181–186. https://doi.org/10.1038/nature02100
Briat JF, Rouached H, Tissot N, Gaymard F, Dubos C (2015) Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front Plant Sci 6:290. https://doi.org/10.3389/fpls.2015.00290
Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6(9):e1001102. https://doi.org/10.1371/journal.pgen.1001102
Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62:185–206. https://doi.org/10.1146/annurev-arplant-042110-103849
Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC (2016) PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res 44:D1154–D1160. https://doi.org/10.1093/nar/gkv1035
Chung BY, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5′ UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120. https://doi.org/10.1186/1471-2164-7-120
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133(2):462–469. https://doi.org/10.1104/pp.103.027979
Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107(1):207–213. https://doi.org/10.1104/pp.107.1.207
Devaux P, Adamski P, Surma M (1992) Inheritance of seed set in crosses of spring barley and Hordeum bulbosum L. Crop Sci 32(1):269–271. https://doi.org/10.2135/cropsci1992.0011183X003200010054x
Frank S (2017) Die Cysteinpeptidase HvPAP14 der Gerste und ihre Rolle beim Abbau plastidärer Proteine. PhD thesis, Kiel University. Retrieved from https://macau.uni-kiel.de/receive/dissertation_diss_00020524
Gallegos JE, Rose AB (2017) Intron DNA sequences can be more important than the proximal promoter in determining the site of transcript initiation. Plant Cell 29(4):843–853. https://doi.org/10.1105/tpc.17.00020
German MA, Luo S, Schroth G, Meyers BC, Green PJ (2009) Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat Protoc 4:356–362. https://doi.org/10.1038/nprot.2009.8
Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K (2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4:1566. https://doi.org/10.1038/ncomms2542
Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, Mao C, Yi K, Wu P, Mo X (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168:1762–1776. https://doi.org/10.1104/pp.15.00736
Hackenberg M, Shi BJ, Gustafson P, Langridge P (2013) Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol 13:214. https://doi.org/10.1186/1471-2229-13-214
Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132(2):578–596. https://doi.org/10.1104/pp.103.020941
Heffer P, Prud’homme M (2013) Nutrients as limited resources: global trends in fertilizer production and use. In: Rengel Z (ed) Improving water and nutrient-use efficiency in food production systems. Wiley, Hoboken, NJ, pp 57–78
Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer-like proteins and HYL1/DRB- family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57(2):173–188. https://doi.org/10.1007/s11103-004-6853-5
Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y, Huang X, Yu F, Kang S, Wang Y, Xie J, Cao S, Zhang L, Wang Y, Xie Q, Kopriva S, Chu C (2019) Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants 5:401–413. https://doi.org/10.1038/s41477-019-0384-1
Huang CY, Shirley N, Genc Y, Shi B, Langridge P (2011) Phosphate utilization efficiency correlates with expression of low-affinity phosphate transporters and noncoding RNA, IPS1, in barley. Plant Physiol 156(3):1217–1229. https://doi.org/10.1104/pp.111.178459
Ishizaki K, Nonomura M, Kato H, Yamato KT, Kohchi T (2012) Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J Plant Res 125(5):643–651. https://doi.org/10.1007/s10265-012-0477-7
Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and MicroRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet 7(3):e1002021. https://doi.org/10.1371/journal.pgen.1002021
Karthikeyan AS, Ballachanda DN, Raghothama KG (2009) Promoter deletion analysis elucidates the role of cis elements and 5′UTR intron in spatiotemporal regulation of AtPht1;4 expression in Arabidopsis. Physiol Plant 136(1):10–18. https://doi.org/10.1111/j.1399-3054.2009.01207.x
Khan GA, Bouraine S, Wege S, Li Y, de Carbonnel M, Berthomieu P, Poirier Y, Rouached H (2014) Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J Exp Bot 65(3):871–884. https://doi.org/10.1093/jxb/ert444
Kim W, Ahn HJ, Chiou TJ, Ahn JH (2011) The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol Cells 32(1):83–88. https://doi.org/10.1007/s10059-011-1043-1
Knop K, Stepien A, Barciszewska-Pacak M, Taube M, Bielewicz D, Michalak M, Borst JW, Jarmolowski A, Szweykowska-Kulinska Z (2017) Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res 45:2757–2775. https://doi.org/10.1093/nar/gkw895
Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15(20):8125–8148. https://doi.org/10.1093/nar/15.20.8125
Kuo HF, Chiou TJ (2011) The role of microRNAs in phosphorus deficiency signaling. Plant Physiol 156(3):1016–1024. https://doi.org/10.1104/pp.111.175265
Kurihara Y, Takashi Y, Watanabe Y (2006) The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12(2):206–212. https://doi.org/10.1261/rna.2146906.2
Lagrange T, Franzetti B, Axelos M, Mache R, Lerbs-Mache S (1993) Structure and expression of the nuclear gene coding for the chloroplast ribosomal protein L21: developmental regulation of a housekeeping gene by alternative promoters. Mol Cell Biol 13(4):2614–2622. https://doi.org/10.1128/mcb.13.4.2614
Li LH, Guo N, Wu ZY, Zhao JM, Sun JT, Wang XT, Xing H (2015) P1BS, a conserved motif involved in tolerance to phosphate starvation in soybean. Genet Mol Res 14(3):9384–9394. https://doi.org/10.4238/2015.August.14.2
Liao L, Ning G, Liu C, Zhang W, Bao M (2013) The intron from the 5′-UTR of the FBP11 gene in petunia displays promoter- and enhancer-like functions. Sci Hortic (Amsterdam) 154:96–101. https://doi.org/10.1016/j.scienta.2013.02.009
Liu TY, Huang TK, Tseng CY, Lai YS, Lin SI, Lin WY, Chen JW, Chiou TJ (2012) PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24(5):2168–2183. https://doi.org/10.1105/tpc.112.096636
Liu Y, Xie Y, Wang H, Ma X, Yao W, Wang H (2017) Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 29(9):2269–2284. https://doi.org/10.1105/tpc.17.00268
Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R (2008) Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Genet Genom 279(6):563–572. https://doi.org/10.1007/s00438-008-0333-6
Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, Wu P (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26(4):1586–1597. https://doi.org/10.1105/tpc.114.123208
Lynch JP (2011) Root phenes for enhanced soil exploration and phosphorus acquisition: tools for future crops. Plant Physiol 156(3):1041–1049. https://doi.org/10.1104/pp.111.175414
Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J (2000) Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J 24(5):559–567. https://doi.org/10.1046/j.1365-313X.2000.00893.x
Melkonyan H, Hofmann HA, Nacken W, Sorg C, Klempt M (1998) The gene encoding the myeloid-related protein 14 (MRP14), a calcium-binding protein expressed in granulocytes and monocytes, contains a potent enhancer element in the first intron. J Biol Chem 273(41):27026–27032. https://doi.org/10.1074/jbc.273.41.27026
Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102(21):7760–7765. https://doi.org/10.1073/pnas.0500778102
Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19(4):1403–1414. https://doi.org/10.1105/tpc.106.048397
Morton T, Petricka J, Corcoran DL, Li S, Winter CM, Carda A, Benfey PN, Ohler U, Megraw M (2014) Paired-end analysis of transcription start sites in Arabidopsis reveals plant-specific promoter signatures. Plant Cell 26(7):2746–2760. https://doi.org/10.1105/tpc.114.125617
Mukatira UT, Liu C, Varadarajan DK, Raghothama KG (2001) Negative regulation of phosphate starvation-induced genes. Plant Physiol 127(4):1854–1862. https://doi.org/10.1104/pp.010876
Muller R, Morant M, Jarmer H, Nilsson L, Nielsen TH (2007) Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol 143(1):156–171. https://doi.org/10.1104/pp.106.090167
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170. https://doi.org/10.3389/fpls.2014.00170
Pacak A, Barciszewska-Pacak M, Swida-Barteczka A, Kruszka K, Sega P, Milanowska K, Jakobsen I, Jarmolowski A, Szweykowska-Kulinska Z (2016) Heat stress affects Pi-related genes expression and inorganic phosphate deposition/accumulation in barley. Front Plant Sci 7:926. https://doi.org/10.3389/fpls.2016.00926
Park BS, Seo JS, Chua NH (2014) NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26(1):454–464. https://doi.org/10.1105/tpc.113.120311
Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39(13):5328–5337. https://doi.org/10.1093/nar/gkr043
Pegler JL, Oultram JMJ, Grof CPL, Eamens AL (2019) DRB1, DRB2 and DRB4 are required for appropriate regulation of the microRNA399/PHOSPHATE2 expression module in Arabidopsis thaliana. Plants (Basel) 8(5):124. https://doi.org/10.3390/plants8050124
Pesole G, Liuni S, Grillo G, Saccone C (1997) Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene 205(1–2):95–102. https://doi.org/10.1016/s0378-1119(97)00407-1
Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276(1–2):73–81. https://doi.org/10.1016/s0378-1119(01)00674-6
Piechulla B, Merforth N, Rudolph B (1998) Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol Biol 38(4):655–662. https://doi.org/10.1023/A:1006094015513
Rose AB, Emami S, Bradnam K, Korf I (2011) Evidence for a DNA-based mechanism of intron-mediated enhancement. Front Plant Sci 2:98. https://doi.org/10.3389/fpls.2011.00098
Ruan W, Guo M, Cai L, Hu H, Li C, Liu Y, Wu Z, Mao C, Yi K, Wu P, Mo X (2015) Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol Biol 87(4–5):429–440. https://doi.org/10.1007/s11103-015-0289-y
Ruan W, Guo M, Wu P, Yi K (2017) Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93(3):327–340. https://doi.org/10.1007/s11103-016-0564-6
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15(16):2122–2133. https://doi.org/10.1101/gad.204401
Schünmann PH, Richardson AE, Smith FW, Delhaize E (2004a) Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J Exp Bot 55(398):855–865. https://doi.org/10.1093/jxb/erh103
Schünmann PH, Richardson AE, Vickers CE, Delhaize E (2004b) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136(4):4205–4214. https://doi.org/10.1104/pp.104.045823
Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25(11):4285–4304. https://doi.org/10.1105/tpc.113.117325
Smoczynska A, Sega P, Stepien A, Knop K, Jarmolowski A, Pacak A, Szweykowska-Kulinska Z (2019) miRNA detection by stem-loop RT-qPCR in studying microRNA biogenesis and microRNA responsiveness to abiotic stresses. Methods Mol Biol 1932:131–150. https://doi.org/10.1007/978-1-4939-9042-9_10
Sobkowiak L, Bielewicz D, Malecka EM, Jakobsen I, Albrechtsen M, Szweykowska-Kulinska Z, Pacak A (2012) The role of the P1BS element containing promoter-driven genes in Pi transport and homeostasis in plants. Front Plant Sci 3:58. https://doi.org/10.3389/fpls.2012.00058
Sonenberg N (1994) mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr Opin Genet Dev 4(2):310–315. https://doi.org/10.1016/s0959-437x(05)80059-0
Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170(1):499–514. https://doi.org/10.1104/pp.15.01336
Tang Z, Sadka A, Morishige DT, Mullet JE (2001) Homeodomain leucine zipper proteins bind to the phosphate response domain of the soybean VspB tripartite promoter. Plant Physiol 125(2):797–809. https://doi.org/10.1104/pp.125.2.797
Thody J, Folkes L, Medina-Calzada Z, Xu P, Dalmay T, Moulton V (2018) PAREsnip2: a tool for high-throughput prediction of small RNA targets from degradome sequencing data using configurable targeting rules. Nucleic Acids Res 46(17):8730–8739. https://doi.org/10.1093/nar/gky609
Thum KE, Kim M, Morishige DT, Eibl C, Koop HU, Mullet JE (2001) Analysis of barley chloroplast psbD light-responsive promoter elements in transplastomic tobacco. Plant Mol Biol 47(3):353–366. https://doi.org/10.1023/A:1011616400264
Todd CD, Zeng P, Huete AM, Hoyos ME, Polacco JC (2004) Transcripts of MYB-like genes respond to phosphorous and nitrogen deprivation in Arabidopsis. Planta 219(6):1003–1009. https://doi.org/10.1007/s00425-004-1305-7
Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthélémy J, Palauqui JC (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20:1494–1503. https://doi.org/10.1105/tpc.107.056069
Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157(3):423–447. https://doi.org/10.1046/j.1469-8137.2003.00695.x
Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H (2012) Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol 196(1):139–148. https://doi.org/10.1111/j.1469-8137.2012.04227.x
Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, Tong Y, Li Z (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot 111(6):1139–1153. https://doi.org/10.1093/aob/mct080
Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K (1999) Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci USA 96(26):15336–15341. https://doi.org/10.1073/pnas.96.26.15336
Yang XJ, Finnegan PM (2010) Regulation of phosphate starvation responses in higher plants. Ann Bot 105(4):513–526. https://doi.org/10.1093/aob/mcq015
Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146(4):1673–1686. https://doi.org/10.1104/pp.107.111443
Acknowledgements
The authors wish to thank Dr. Iver Jakobsen (University of Copenhagen, Copenhagen, Denmark) for providing low-P soil and Prof Tzyy-Jen Chiou (Agricultural Biotechnology Research Center, Academia Sinica, Taiwan) for the construct containing the AtPHO2 promoter and the 5′-UTR. We thank Michał Taube and Przemysław Wieczorek for their help with the protein overexpression and purification protocols and Mateusz Bajczyk for all his advice.
Funding
This work was funded by the National Science Centre, Poland, on the basis of DEC-2013/11/B/NZ9/01761, UMO-2016/23/B/NZ9/00857, and UMO-2015/19/N/NZ9/00218 and by KNOW RNA Research Centre in Poznan 01/KNOW2/2014.
Author information
Authors and Affiliations
Contributions
PS performed most of the experiments and wrote the manuscript under the supervision of AP. AP prepared the degradome and transcriptome libraries. LS performed the protein subcellular localization. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries (AtNSR1 (AAF05867), MYB-1 (AK373855), MYB-2/APL (AK371403), MADS57 (AK363243)) and the Ensembl Plants database for the barley genome (PHR1 (HORVU4Hr1G051080.5), PHR2 (HORVU4Hr1G051080.1), MYB-2/APL (HORVU6Hr1G031470), PHO2 (HORVU1Hr1G085570.2)).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
List of proteins obtained from Y1H experiments. Supplementary Table 2 Synthetic oligonucleotides used for EMSA experiments. Supplementary Table 3 List of selected proteins used for the phylogenetic analysis of the APL TF. Supplementary Table 4 Primers used in this study. Supplementary Table 5 Y1H “bait” sequences derived from PHO2 gene fragments used for screening. (DOCX 29 kb)
Supplementary Fig. S1
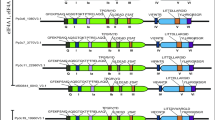
The graph showing all six PHO2 transcript variants from the Ensembl Plants database and RNA-Seq results. There are six PHO2 protein-coding transcripts that are present in the barley Ensembl Plants database: HORVU1Hr1G085570.1 (544 aa), HORVU1Hr1G085570.2 (847 aa), HORVU1Hr1G085570.3 (445 aa), HORVU1Hr1G085570.4 (847 aa), HORVU1Hr1G085570.5 (606 aa), and HORVU1Hr1G085570.6 (491 aa). The lower panel displays the cDNA sequences of the first 180 bp from the HORVU1Hr1G085570.1 variant and the last 26 bp of the HORVU1Hr1G085570.2 variant to show the 5′ (blue arrow) and 3′ (red arrows) ends obtained in our RNA-Seq analysis. The white boxes indicate the untranslated region, and the gray boxes indicate the coding region. Scale bar = 1000 bp. Supplementary Fig. S2 The genomic sequence of the barley PHO2 5′-UTR region used in this study. The validated genomic sequence of the barley PHO2 5′-UTR region (2742 bp) that was cloned and sequenced from the Morex genotype (Ensembl Plants database: chr1H:535891650:535894391:1). Denoted are the cis-regulatory motifs that were identified in this study, in addition to six miR399 potential cleavage sites. The yellow boxes indicate the locations of exons. The black star indicates the additional exon within the 5′-UTR that is present only in barley PHO2 isoforms 5 and 6. Supplementary Fig. S3 The P1BS motif is evolutionarily conserved and present in various PSI genes in barley. The genomic localization of P1BS motifs within regulatory sequences of the barley Pi-starvation-responsive genes PHO2, MIR399c, IPS1, RNS1, and PHT1;1. The Arabidopsis thaliana (AtPHO2) and Nicotiana benthamiana (NbPHO2) PHO2 gene orthologs were used as a reference to show the P1BS motif positions within barley PHO2 regulatory sequences relative to those within the other plant species used in this study. The gray box depicts the 5′-UTR; the white box depicts the promoter; the red line indicates the position of the P1BS motif. Scale bar = 1000 bp (left). Comparison of the sequences of all P1BS motifs that are present in the left panel (right). The yellow box indicates the specific nucleotide within the motif consensus; the consensus match connects barley PHO2 motifs with P1BS motifs that are the same but present within the regulatory sequences of other genes. Supplementary Fig. S4 The barley PHO2 transcript is cleaved within its 5′-UTR. The red vertical line shows the cleavage position directed by miR399; the cleavage position 1203 is within exon No. 2 in the 5′-UTR of the PHO2 transcript (HORVU1Hr1G085570.2, length of 4347 nt). The black vertical lines on the graph show the positions within the PHO2 cDNA to which 20 nt degradome fragments (reads) were mapped. The number of such reads (fragment abundance) is depicted by the height of the red and black lines. Below the graph, the structure of the PHO2 transcript is presented. The white boxes denote UTRs, the gray boxes denote CDSs, and the dotted vertical lines denote cleavage sites within the 5′-UTR. Supplementary Fig. S5 The multiple sequence alignment for PHR-like TFs in which the AtNSR1 protein sequence was used as a query. On the basis of the results published by Todd and his group in 2004, we selected two MYB-like TFs (temporarily named MYB-1 and MYB-2) that exhibited the highest homology to the AtNSR1 protein (At3g04030) and analyzed them in a Y1H screening assay. The orange box indicates the SANT/MYB protein domain, and the green box indicates the MYB-CC domain. Supplementary Fig. S6 The barley TFs PHR1 and APL interact with the second exon “bait” fragment originating from the PHO2 5′-UTR in yeast cells. The structure of the barley PHO2 gene with marked a Y1H “bait” fragment. The pPHO2_5 fragment is 27 bp in length and includes the P1BS.2 motif. The blue triangle indicates positions 447916090 (+) and 447916332 (+) on barley chromosome 1; the two TSSs of the PHO2 transcripts were identified using 5′RLM-RACE. The lower panel contains images of the growing colonies of the tested Y1HGold yeast strain having the “bait” fragment from the PHO2 5′-UTR second exon. The barley MADS57 TF was used as a non-binding negative control. Supplementary Fig. S7 The first intron originating from the barley PHO2 5′-UTR contains signals that help increase gene expression. The distribution of IMEter scores for either (A) all nine PHO2-related introns (the first two introns are located within 5′-UTR region) or (B) seven fragments approximately 200 bp in length chopped from the first intron of the PHO2 5′-UTR. Each ~200 bp fragment is designated with an additional prefix from p1 to p7, where the p1 fragment is located on the first intron 5′-end, and p7 is located on its 3′-end. The plots show percentage scores, which were calculated on the basis of the results from three monocotyledonous plant species: Brachypodium distachyon, Oryza sativa, and Zea mays. Supplementary Fig. S8PHO2, PHR1, and APL transcript coverage. RNA-Seq paired reads were mapped to the PHO2 (HORVU1Hr1G085570.2, Ensembl Plants database), PHR1 (HORVU4Hr1G051080.5), and APL (HORVU6Hr1G031470.1) transcripts. Blue indicates the reverse paired reads, green indicates forward unpaired reads, and red indicates reverse unpaired reads; the black vertical line denotes the ATG start codon. Below the graph, the structures of the transcript are presented. The white boxes denote UTRs, and the gray boxes denote CDSs. Supplementary Fig. S9 The new protein identified in Y1H screening has two highly conserved protein domains and is likely related to the APL TFs. Phylogenetic analysis was carried out using the neighbor-joining method in CLC Main Workbench software (QIAGEN). To select proteins for the cladogram, we used the APL protein sequence as a query, and proteins showing more than 60% homology were chosen for analysis. Additionally, the pool of chosen proteins was enriched with several well-characterized plant PHR-like proteins. Each bootstrap value was calculated with 100 replications. All accession numbers of the protein sequences are listed in Table S3. The black arrow shows the APL position on the tree. The lower panel presents the protein structures of the barley APL, PHR1, and PHR2 proteins. The APL protein contains 368 aa residues (39.85 kDa) that comprise two domains: a SANT/MYB domain (orange box, PF00249) at aa residues 45–93 and a MYB-CC-type LHEQLE-containing domain (blue box, PF14379) at aa residues 139–193. The PHR1 protein contains 451 aa residues (49.48 kDa), and the same domains are present at aa residues 233–284 and residues 314–360; however, the PHR2 protein contains 438 aa residues (47.17 kDa), and the domains are located at residues 256–307 and residues 340–385, respectively. Scale bar = 100 aa. Supplementary Fig. S10 The cellular localization of PHR1–eGFP fusion proteins in barley protoplasts. Exclusive images that show the localization of recombinant proteins transiently expressed in protoplasts isolated from 6-day-old barley leaves. Slight expression of PHR1–eGFP proteins in the cytoplasm was observed in one out of every five cells. The protoplasts shown here were incubated overnight in standard W5 buffer (without extra KH2PO4, a source of Pi). Microscopic analyses were repeated three times, and similar patterns were imaged. Scale bars = 20 µm. (PDF 12523 kb)
Rights and permissions
About this article
Cite this article
Sega, P., Kruszka, K., Szewc, Ł. et al. Identification of transcription factors that bind to the 5′-UTR of the barley PHO2 gene. Plant Mol Biol 102, 73–88 (2020). https://doi.org/10.1007/s11103-019-00932-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00932-9