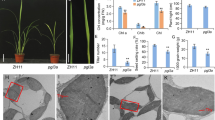
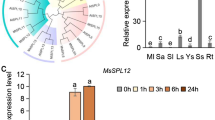
Abstract
Damaged proteins containing abnormal isoaspartyl (isoAsp) accumulate as seeds age and the abnormality is thought to undermine seed vigor. Protein-l-isoaspartyl methyltransferase (PIMT) is involved in isoAsp-containing protein repair. Two PIMT genes from rice (Oryza sativa L.), designated as OsPIMT1 and OsPIMT2, were isolated and investigated for their roles. The results indicated that OsPIMT2 was mainly present in green tissues, but OsPIMT1 largely accumulated in embryos. Confocal visualization of the transient expression of OsPIMTs showed that OsPIMT2 was localized in the chloroplast and nucleus, whereas OsPIMT1 was predominately found in the cytosol. Artificial aging results highlighted the sensitivity of the seeds of OsPIMT1 mutant line when subjected to accelerated aging. Overexpression of OsPIMT1 in transgenic seeds reduced the accumulation of isoAsp-containing protein in embryos, and increased embryo viability. The germination percentage of transgenic seeds overexpressing OsPIMT1 increased 9–15 % compared to the WT seeds after 21-day of artificial aging, whereas seeds from the OsPIMT1 RNAi lines overaccumulated isoAsp in embryos and experienced rapid loss of seed germinability. Taken together, these data strongly indicated that OsPIMT1-related seed longevity improvement is probably due to the repair of detrimental isoAsp-containing proteins that over accumulate in embryos when subjected to accelerated aging.









Similar content being viewed by others
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645. doi:10.1074/jbc.M605895200
Baranowskij N, Frohberg C, Prat S, Willmitzer L (1994) A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J 13:5383–5392
Bartels D, Souer E (2004) Molecular responses of higher plants to dehydration. In: Hirt H, Shinozaki K (eds) Plant responses to abiotic stress, vol 4., Topics in current geneticsSpringer, Berlin Heidelberg, pp 9–38
Bernal-Lugo I, Rodriguez M, Gavilanes-Ruiz M, Hamabata A (1999) Reduced aleurone α-amylase production in aged wheat seeds is accompanied by lower levels of high-pI α-amylase transcripts and reduced response to gibberellic acid. J Exp Bot 50:311–317. doi:10.1093/jxb/50.332.311
Chatelain E, Satour P, Laugier E, Ly VuB, Payet N, Rey P, Montrichard F (2013) Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proc Natl Acad Sci 110(9):3633–3638
Chen T, Nayak N, Majee SM, Lowenson J, Schafermeyer KR, Eliopoulos AC, Lloyd TD, Dinkins R, Perry SE, Forsthoefel NR (2010) Substrates of the Arabidopsis thaliana protein isoaspartyl methyltransferase 1 identified using phage display and biopanning. J Biol Chem 285:37281–37292
Chen H, Chu P, Zhou Y, Li Y, Liu J, Ding Y, Tsang EWT, Jiang L, Wu K, Huang S (2012) Overexpression of AtOGG1, a DNA glycosylase/AP lyase, enhances seed longevity and abiotic stress tolerance in Arabidopsis. J Exp Bot 63:4107–4121
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054. doi:10.1101/gad.1077503
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55:225–236. doi:10.1093/jxb/erh005
Clarke S (2003) Aging as war between chemical and biochemical processes: protein methylation and the recognition of age-damaged proteins for repair. Ageing Res Rev 2:263–285
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32:40–52
Das G, Sen-Mandi S (1988) Root formation in deteriorated (aged) wheat embryos. Plant Physiol 88:983–986. doi:10.1104/pp.88.4.983
Debeaujon I, Leon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122(2):403–414
Dinkins RD, Majee SM, Nayak NR, Martin D, Xu Q, Belcastro MP, Houtz RL, Beach CM, Downie AB (2008) Changing transcriptional initiation sites and alternative 5′-and 3′-splice site selection of the first intron deploys Arabidopsis PROTEIN ISOASPARTYL METHYLTRANSFERASE2 variants to different subcellular compartments. Plant J 55:1–13
Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell Online 21:972–984
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Finkelstein R (2010) The role of hormones during seed development and germination. In: Davies P (ed) Plant hormones. Springer, Netherlands, pp 549–573. doi:10.1007/978-1-4020-2686-7_24
Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P (2004) cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell Online 16:1077–1090. doi:10.1105/tpc.019729
Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier C, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587
Kagan RM, McFadden HJ, McFadden PN, O’Connor C, Clarke S (1997a) Molecular phylogenetics of a protein repair methyltransferase. Comp Biochem Physiol B: Biochem Mol Biol 117:379–385
Kagan RM, Niewmierzycka A, Clarke S (1997b) Targeted gene disruption of the Caenorhabditis elegans l-isoaspartyl protein repair methyltransferase impairs survival of dauer stage nematodes. Arch Biochem Biophys 348:320–328. doi:10.1006/abbi.1997.0362
Kaplan B, Davydov O, Knight H, Galon Y, Knight MR, Fluhr R, Fromm H (2006) Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell Online 18:2733–2748. doi:10.1105/tpc.106.042713
Kim E, Lowenson JD, MacLaren DC, Clarke S, Young SG (1997) Deficiency of a protein-repair enzyme results in the accumulation of altered proteins, retardation of growth, and fatal seizures in mice. Proc Natl Acad Sci 94:6132
Lafitte H, Yongsheng G, Yan S, Li Z-K (2007) Whole plant responses, key processes, and adaptation to drought stress: the case of rice. J Exp Bot 58:169–175. doi:10.1093/jxb/erl101
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155–3175. doi:10.1105/tpc.105.035568
Li C, Clarke S (1992) A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci 89:9885–9889
Li X, Bai H, Wang X, Li L, Cao Y, Wei J, Liu Y, Liu L, Gong X, Wu L (2011) Identification and validation of rice reference proteins for western blotting. J Exp Bot 62:4763–4772
Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229:605–615
Mudgett MB, Clarke S (1993) Characterization of plant l-isoaspartyl methyltransferases that may be involved in seed survival: purification, cloning, and sequence analysis of the wheat germ enzyme. Biochemistry 32:11100–11111
Mudgett MB, Clarke S (1994) Hormonal and environmental responsiveness of a developmentally regulated protein repair l-isoaspartyl methyltransferase in wheat. J Biol Chem 269:25605–25612
Mudgett M, Clarke S (1996) A distinctly regulated protein repair l-isoaspartylmethyltransferase from Arabidopsis thaliana. Plant Mol Biol 30:723–737. doi:10.1007/BF00019007
Mudgett MB, Lowenson JD, Clarke S (1997) Protein repair l-isoaspartyl methyltransferase in plants (phylogenetic distribution and the accumulation of substrate proteins in aged barley seeds). Plant Physiol 115:1481–1489. doi:10.1104/pp.115.4.1481
Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41:697–709. doi:10.1111/j.1365-313X.2005.02337.x
Niu X (2002) Maize ABI4 binds coupling element1 in abscisic acid and sugar response genes. Plant Cell Online 14:2565–2575. doi:10.1105/tpc.003400
Nonogaki H (2006) Seed germination—The biochemical and molecular mechanisms. Breed Sci 56:93–105. doi:10.1270/jsbbs.56.93
Oge L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P (2008) Protein repair l-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 20:3022–3037. doi:10.1105/tpc.108.058479
Petruzzelli L, Taranto G (1989) Wheat aging: the contribution of embryonic and non-embryonic lesions to loss of seed viability. Physiol Plant 76:289–294. doi:10.1111/j.1399-3054.1989.tb06193.x
Rajjou L, Debeaujon I (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. CR Biol 331:796–805
Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63:507–533
Reddy ASN, Ali GS, Celesnik H, Day IS (2011) Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell Online 23:2010–2032. doi:10.1105/tpc.111.084988
Rock C, Quatrano R (1995) The role of hormones during seed development. In: Davies P (ed) Plant hormones. Springer, Netherlands, pp 671–697. doi:10.1007/978-94-011-0473-9_31
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi:10.1038/nprot.2008.73
Schurter BT, Aswad DW (2000) Analysis of isoaspartate in peptides and proteins without the use of radioisotopes. Anal Biochem 282:227–231. doi:10.1006/abio.2000.4601
Shen-Miller J (2002) Sacred lotus, the long-living fruits of China Antique. Seed Sci Res 12:131–143. doi:10.1079/SSR2002112
Shirsat A, Wilford N, Croy R, Boulter D (1989) Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet: MGG 215:326–331
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang R-S, Zheng J-C, Jin Z-Q, Zhang D-D, Huang Y-H, Chen L-G (2008) Possible correlation between high temperature-induced floret sterility and endogenous levels of IAA, GAs and ABA in rice (Oryza sativa L.). Plant Growth Regul 54:37–43. doi:10.1007/s10725-007-9225-8
Thapar N, Kim A-K, Clarke S (2001) Distinct patterns of expression but similar biochemical properties of protein l-isoaspartyl methyltransferase in higher plants. Plant Physiol 125:1023–1035
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539. doi:10.1105/tpc.5.11.1529
Verma P, Singh A, Kaur H, Majee M (2010) PROTEIN l--ISOASPARTYL METHYLTRANSFERASE1 (CaPIMT1) from chickpea mitigates oxidative stress-induced growth inhibition of Escherichia coli. Planta 231:329–336
Verma P, Kaur H, Petla BP, Rao V, Saxena SC, Majee M (2013) PROTEIN l-ISOASPARTYL METHYLTRANSFERASE2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol 161:1141–1157
Villa ST, Xu Q, Downie AB, Clarke SG (2006) Arabidopsis protein repair l-isoaspartyl methyltransferases: predominant activities at lethal temperatures. Physiol Plant 128:581–592
Visick JE, Cai H, Clarke S (1998) The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol 180:2623–2629
Wang Z, Chen C, Xu Y, Jiang R, Han Y, Xu Z, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22:409–417. doi:10.1007/BF02772683
Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, West CE (2010) A plant DNA ligase is an important determinant of seed longevity. Plant J 63:848–860. doi:10.1111/j.1365-313X.2010.04285.x
Xu Q, Belcastro MP, Villa ST, Dinkins RD, Clarke SG, Downie AB (2004) A second protein l-isoaspartyl methyltransferase gene in Arabidopsis produces two transcripts whose products are sequestered in the nucleus. Plant Physiol 136:2652
Yang T, Poovaiah BW (2002) A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277:45049–45058. doi:10.1074/jbc.M207941200
Yoshida S, Forno D, Cock J, Gomez K (eds) (1976) Routine procedure for growing rice plants in culture solution. In: Laboratory manual for physiological studies of rice. The International Rice Research Institute, 3rd edn. Los Baños, Laguna, Philippines, pp 61–66
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685. doi:10.1111/j.1365-313X.2009.04092.x
Zeng D-L, Qian Q, Yasukumi K (2002) Study on storability and morphological index in rice (Oryza sativa L.) under artificial ageing. Acta Agron Sin 28:551–554
Zhang W, Ruan J, T-hD Ho, You Y, Yu T, Quatrano RS (2005) Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics 21:3074–3081
Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, Wang J, Wang H (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. doi:10.1186/1746-4811-7-30
Zhou Z, Robards K, Helliwell S, Blanchard C (2002) Ageing of stored rice: changes in chemical and physical attributes. J Cereal Sci 35:65–78
Acknowledgments
We thank Prof. Jiayang Li at Chinese Academy of Agricultural Sciences, Dr. Zhen Li at Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for technical support of subcellular localization. We thank National key laboratory of Crop genetics improvement, Huazhong Agricultural University for the mutant of OsPIMT1 derived from variety ZH11. We also thank Prof. Kang Chong of Institute of Botany, Chinese Academy of Sciences for the RNAi vector. The research is supported by grants from The Hi-Tech Research and Development (863) Program of China (2014AA10A603, 2014AA10A604), The National Major Projects of Cultivated Transgenic New Crop Varieties Foundation of China (2008ZX001-006, 2011ZX001-006), The National Natural Science Foundation of China (31401471), The Special Foundation of Non⁃Profit Research Institutes of Fujian Province (2014R1021-8), and The Science Fund for Distinguished Young Scholars of Fujian Provincial Academy of Agricultural Sciences (2014JQ-3).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Accession numbers Sequence data from this article can be found in GenBank/EMBL data bases under the accession numbers listed below. The newly obtained cDNA sequences of Os08g0557000/OsPIMT1 and Os04g0481400/OsPIMT2 were deposited to GenBank under accession number KM527502 and KM675736, respectively. Accession Numbers used in sequence blast and phylogenetic analysis are in Supplemental Table S2.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure S1
Hydrophobicity analysis and targeting prediction of OsPIMT2 (TIFF 2288 kb)
Supplemental Figure S2
Prokaryotic expression analysis of OsPIMT1 and OsPIMT2 (TIFF 974 kb)
Supplemental Figure S3
Expression analysis of OsPIMT1 and OsPIMT2 under salicylic acid and jasmonic acid treatment (TIFF 883 kb)
Supplemental Figure S4
Verification of pimt1-1 T-DNA insertion and screening of homozygous pimt1-1 mutant plants (TIFF 231 kb)
Supplemental Figure S5
Overall view of germination performance of OsPIMT1 transgenic lines and WT after aging treatment (TIFF 5961 kb)
Supplemental Table S1
List of Primers used in this study (DOC 55 kb)
Supplemental Table S2
Accession numbers used in sequence blast and phylogenetic analysis (DOC 38 kb)
Supplemental Table S3
List of cis-regulatory elements in the 750-bp promoter regions of OsPIMT1 and OsPIMT2 (DOC 78 kb)
Rights and permissions
About this article
Cite this article
Wei, Y., Xu, H., Diao, L. et al. Protein repair l -isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol Biol 89, 475–492 (2015). https://doi.org/10.1007/s11103-015-0383-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0383-1