Abstract
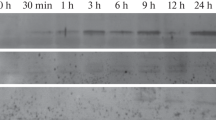
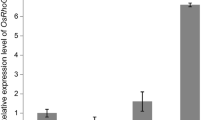
A Glycine max gene encoding a putative protein similar to hypersensitive-induced response proteins (HIR) was identified as a gene with preferred expressions in flowers and developing seeds by whole transcriptome gene expression profiling. Its promoter gm-hir1 was cloned and revealed to strongly express a fluorescence reporter gene primarily in integuments, anther tapetum, and seed coat with unique tissue-specificity. Expression in the inner integument was apparent prior to pollination, while expression in the outer integument started to develop from the micropylar end outward as the embryo matured. A 5′-deletion study showed that the promoter can be truncated to 600 bp long relative to the translation start site without affecting expression. A positive regulatory element was identified between 600 and 481 bp that controls expression in the inner integument, with no noticeable effect on expression in the outer integument or tapetum. Additionally, removal of the 5′UTR intron had no effect on levels or location of gm-hir1 expression while truncation to 370 bp resulted in a complete loss of expression suggesting that elements controlling both the outer integument and tapetum expression are located within the 481–370 bp region.






Similar content being viewed by others
References
Batchelor AK, Boutilier K, Miller SS, Hattori J, Bowman LA, Hu M, Lantin S, Johnson DA, Miki BLA (2002) SCB1, a BURP-domain protein gene, from developing soybean seed coats. Planta 215:523–532
Brownman DT, Hoegg MB, Robbins SM (2007) The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 17:394–402
Chamberlin MA, Horner HT, Palmer RG (1994) Early endosperm, embryo, and ovule development in Glycine max (L.) Merr. Int J Plant Sci 155:421–436
Falco SC, Li Z (2007) S-adenosyl-l-methionine synthetase promoter and its use in expression of transgenic genes in plants. US patent 7,217,858
Gui M, Liu W (2014) Programmed cell death during floral nectary senescence in Ipomoea purpurea. Protoplasma 251:677–685
Hakozaki H, Endo M, Masuko H, Park JI, Ito H, Uchida M, Kamada M, Takahashi H, Higashitani A, Watanabe M (2004) Cloning and expression pattern of a novel microspore-specific gene encoding hypersensitive-induced response protein (LjH1R1) from the model legume, Lotus japonicus. Genes Genet Syst 79:307–310
Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28:837–849
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Horner HT, Healy RA, Cervantes-Martinez T, Palmer RG (2003) Floral nectary fine structure and development in Glycine max L. (Fabaceae). Int J Plant Sci 164:675–690
Huang MD, Chen TLL, Huang AHC (2013) Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiol 163:1218–1229
Langhorst MF, Reuter A, Struermer CAO (2005) Scaffolding microdomains and beyond: the function of reggi/flotillin proteins. Cell Mol Life Sci 62:2228–2240
Li Z, Xing A, Moon BP, Burgoyne SA, Guida AD, Liang H, Lee C, Caster CS, Barton JE, Klein TM, Falco SC (2007) A Cre/loxP-mediated self-activating gene excision system to produce marker gene free transgenic soybean plants. Plant Mol Biol 65:329–341
Liu L, Fan X (2013) Tapetum: regulation and role in soropollenin biosynthesis in Arabidopsis. Plant Mol Biol 83:165–175
Liu X, Shangguan Y, Zhu J, Lu Y, Han B (2013) The rice OsLTP6 gene promoter directs anther-specific expression by a combination of positive and negative regulatory elements. Planta 238:845–857
Meister RJ, Williams LA, Monfared MM, Gallagher TL, Kraft EA, Nelson CG, Gasser CS (2004) Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J 37:426–438
Miller SS, Bowman LA, Gijzen M, Miki BLA (1999) Early development of the seed coat of soybean (Glycine max). Ann Bot 84:297–304
Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, Ueno K, Mochizuki A, Tanimoto H, Tsugawa H, Otsuki Y, Ohashi Y (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37:49–59
Mongkolsiriwatana C, Pongtongkam P, Peyachoknagul S (2009) In silico promoter analysis of photoperiod-responsive genes identified by DNA microarray in rice (Oryza sativa L.). Kasetsart J (Nat Sci) 43:164–177
Nadimpalli R, Yalpani N, Johal GS, Simmons CR (2000) Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem 275:29579–29586
Odell JT, Nagy F, Chua N (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Rivera-Milla E, Stuermer CAO, Málaga-Trilli E (2006) Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci 63:343–357
Rose AB (2008) Intron-mediated regulation of gene expression. Curr Top Microbiol 326:277–290
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Simon MK, Williams LA, Brady-Passerini K, Brown RH, Gasser CS (2012) Positive- and negative-acting regulatory elements contribute to the tissue-specific expression of INNER NO OUTER, a YABBY-type transcription factor gene in Arabidopsis. BMC Plant Biol 12:214
Stadler R, Lauterbach C, Sauer N (2005) Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol 139:701–712
Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J (2008) Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453:1239–1243
Wu LM, El-Mezawy A, Shah S (2011) A seed coat outer integument-specific promoter for Brassica napus. Plant Cell Rep 30:75–80
Xing A, Moon BP, Mills KM, Falco SC, Li Z (2010) Revealing frequent alternative polyadenylation and widespread low-level transcription read-through of novel plant transcription terminators. Plant Biotech J 8:772–782
Yamauchi D (2001) A TGACGT motif in the 5′-upstream region of α-amylase gene from Vigna mungo is a cis-element for expression in cotyledons of germinated seeds. Plant Cell Physiol 42:635–641
Yu XM, Zhao WQ, Yang WX, Liu F, Chen JP, Goyer C, Liu DQ (2013) Characterization of a hypersensitive response-induced gene TaHIR3 from wheat leaves infected with leaf rust. Plant Mol Biol Rep 31:314–322
Yu J, Zhang Z, Wei J, Ling Y, Xu W, Su Z (2014) SFGD: a comprehensive platform for mining functional information from soybean transcriptome data and its use in identifying acyl-lipid metabolism pathways. BMC Genom 15:271
Zhou L, Cheung MY, Li MW, Fu Y, Sun Z, Sun SM, Lam HM (2010) Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol 10:290
Acknowledgments
We are grateful to DuPont soybean transformation group for producing transgenic soybean plants, and DuPont core facilities for sequencing, PCR analysis, and plant maintenance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koellhoffer, J.P., Xing, A., Moon, B.P. et al. Tissue-specific expression of a soybean hypersensitive-induced response (HIR) protein gene promoter. Plant Mol Biol 87, 261–271 (2015). https://doi.org/10.1007/s11103-014-0274-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-014-0274-x