Abstract
Background
Venlafaxine (VEN) is a commonly utilized medication for alleviating depression and anxiety disorders. The presence of genetic polymorphisms gives rise to considerable variations in plasma concentrations across different phenotypes. This divergence in phenotypic responses leads to notable differences in both the efficacy and tolerance of the drug.
Purpose
A physiologically based pharmacokinetic (PBPK) model for VEN and its metabolite O-desmethylvenlafaxine (ODV) to predict the impact of CYP2D6 and CYP2C19 gene polymorphisms on VEN pharmacokinetics (PK).
Methods
The parent-metabolite PBPK models for VEN and ODV were developed using PK-Sim® and MoBi®. Leveraging prior research, derived and implemented CYP2D6 and CYP2C19 activity score (AS)-dependent metabolism to simulate exposure in the drug-gene interactions (DGIs) scenarios. The model’s performance was evaluated by comparing predicted and observed values of plasma concentration–time (PCT) curves and PK parameters values.
Results
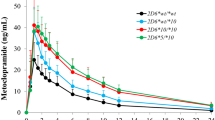
In the base models, 91.1%, 94.8%, and 94.6% of the predicted plasma concentrations for VEN, ODV, and VEN + ODV, respectively, fell within a twofold error range of the corresponding observed concentrations. For DGI scenarios, these values were 81.4% and 85% for VEN and ODV, respectively. Comparing CYP2D6 AS = 2 (normal metabolizers, NM) populations to AS = 0 (poor metabolizers, PM), 0.25, 0.5, 0.75, 1.0 (intermediate metabolizers, IM), 1.25, 1.5 (NM), and 3.0 (ultrarapid metabolizers, UM) populations in CYP2C19 AS = 2.0 group, the predicted DGI AUC0-96 h ratios for VEN were 3.65, 3.09, 2.60, 2.18, 1.84, 1.56, 1.34, 0.61, and for ODV, they were 0.17, 0.35, 0.51, 0.64, 0.75, 0.83, 0.90, 1.11, and the results were similar in other CYP2C19 groups. It should be noted that PK differences in CYP2C19 phenotypes were not similar across different CYP2D6 groups.
Conclusions
In clinical practice, the impact of genotyping on the in vivo disposition process of VEN should be considered to ensure the safety and efficacy of treatment.







Similar content being viewed by others
Data Availability
Upon reasonable request, the corresponding author can provide the available data.
References
Marwaha S, Palmer E, Suppes T, Cons E, Young AH, Upthegrove R. Novel and emerging treatments for major depression. Lancet. 2023;401(10371):141–53.
FDA. Lable-EFFEXOR XR® (venlafaxine extended-release) capsules 2023 [Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/020699s118lbl.pdf.
Zhou Q, Li X, Yang D, Xiong C, Xiong Z. A comprehensive review and meta-analysis of neurological side effects related to second-generation antidepressants in individuals with major depressive disorder. Behav Brain Res. 2023;447:114431.
Fogelman SM, Schmider J, Venkatakrishnan K, von Moltke LL, Harmatz JS, Shader RI, et al. O- and N-demethylation of venlafaxine in vitro by human liver microsomes and by microsomes from cDNA-transfected cells: effect of metabolic inhibitors and SSRI antidepressants. Neuropsychopharmacology. 1999;20(5):480–90.
Liebowitz MR, Tourian KA. Efficacy, safety, and tolerability of desvenlafaxine 50 mg/d for the treatment of major depressive disorder:a systematic review of clinical trials. Prim Care Companion J Clin Psychiatry. 2010;12(3):PCC.09r00845.
Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369(1):23–37.
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–41.
Rüdesheim S, Selzer D, Mürdter T, Igel S, Kerb R, Schwab M, et al. Physiologically based pharmacokinetic modeling to describe the CYP2D6 activity score-dependent metabolism of paroxetine, atomoxetine and risperidone. Pharmaceutics. 2022;14(8):1734.
Gaedigk A, Dinh JC, Jeong H, Prasad B, Leeder JS. Ten years’ experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J Pers Med. 2018;8(2):15.
Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the clinical pharmacogenetics implementation consortium and dutch pharmacogenetics working group. Clin Transl Sci. 2020;13(1):116–24.
Shams ME, Arneth B, Hiemke C, Dragicevic A, Müller MJ, Kaiser R, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther. 2006;31(5):493–502.
Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102(1):37–44.
Yang H, Yang L, Zhong X, Jiang X, Zheng L, Wang L. Physiologically based pharmacokinetic modeling of brivaracetam and its interactions with rifampin based on CYP2C19 phenotypes. Eur J Pharm Sci. 2022;177:106258.
Black JL 3rd, O’Kane DJ, Mrazek DA. The impact of CYP allelic variation on antidepressant metabolism: a review. Expert Opin Drug Metab Toxicol. 2007;3(1):21–31.
Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9(5):442–73.
FDA. The Use of Physiologically Based Pharmacokinetic Analyses — Biopharmaceutics Applications for Oral Drug Product Development, Manufacturing Changes, and Controls 2020 [Available from: https://www.fda.gov/media/142500/download.
Lesser IM, Zisook S, Gaynes BN, Wisniewski SR, Luther JF, Fava M, et al. Effects of race and ethnicity on depression treatment outcomes: the CO-MED trial. Psychiatr Serv. 2011;62(10):1167–79.
Shen C, Shao W, Wang W, Sun H, Wang X, Geng K, et al. Physiologically based pharmacokinetic modeling of levetiracetam to predict the exposure in hepatic and renal impairment and elderly populations. CPT Pharmacometrics Syst Pharmacol. 2023;12(7):1001–15.
Willmann S, Thelen K, Lippert J. Integration of dissolution into physiologically-based pharmacokinetic models III: PK-Sim®. J Pharm Pharmacol. 2012;64(7):997–1007.
McAlpine DE, Biernacka JM, Mrazek DA, O’Kane DJ, Stevens SR, Langman LJ, et al. Effect of cytochrome P450 enzyme polymorphisms on pharmacokinetics of venlafaxine. Ther Drug Monit. 2011;33(1):14–20.
Rüdesheim S, Selzer D, Fuhr U, Schwab M, Lehr T. Physiologically-based pharmacokinetic modeling of dextromethorphan to investigate interindividual variability within CYP2D6 activity score groups. CPT Pharmacometrics Syst Pharmacol. 2022;11(4):494–511.
Jones HM, Chen Y, Gibson C, Heimbach T, Parrott N, Peters SA, et al. Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin Pharmacol Ther. 2015;97(3):247–62.
Sager JE, Yu J, Ragueneau-Majlessi I, Isoherranen N. Physiologically based pharmacokinetic (PBPK) modeling and simulation approaches: a systematic review of published models, applications, and model verification. Drug Metab Dispos. 2015;43(11):1823–37.
Shen C, Liang D, Wang X, Shao W, Geng K, Wang X, et al. Predictive performance and verification of physiologically based pharmacokinetic model of propylthiouracil. Front Pharmacol. 2022;13:1013432.
Hanke N, Frechen S, Moj D, Britz H, Eissing T, Wendl T, et al. PBPK models for CYP3A4 and P-gp DDI prediction: a modeling network of rifampicin, itraconazole, clarithromycin, midazolam, alfentanil, and digoxin. CPT Pharmacometrics Syst Pharmacol. 2018;7(10):647–59.
Zheng L, Yang H, Dallmann A, Jiang X, Wang L, Hu W. Physiologically based pharmacokinetic modeling in pregnant women suggests minor decrease in maternal exposure to olanzapine. Front Pharmacol. 2021;12:793346.
Yim DS, Bae SH, Choi S. Predicting human pharmacokinetics from preclinical data: clearance. Transl Clin Pharmacol. 2021;29(2):78–87.
Rüdesheim S, Wojtyniak JG, Selzer D, Hanke N, Mahfoud F, Schwab M, et al. Physiologically based pharmacokinetic modeling of metoprolol enantiomers and α-hydroxymetoprolol to describe CYP2D6 drug-gene interactions. Pharmaceutics. 2020;12(12):1200.
Guest EJ, Aarons L, Houston JB, Rostami-Hodjegan A, Galetin A. Critique of the two-fold measure of prediction success for ratios: application for the assessment of drug-drug interactions. Drug Metab Dispos. 2011;39(2):170–3.
FDA. DESVENLAFAXINE extended-release tablets 2023 [Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/204150s020lbl.pdf.
Calleja S, Zubiaur P, Ochoa D, Villapalos-García G, Mejia-Abril G, Soria-Chacartegui P, et al. Impact of polymorphisms in CYP and UGT enzymes and ABC and SLCO1B1 transporters on the pharmacokinetics and safety of desvenlafaxine. Front Pharmacol. 2023;14:1110460.
Fukuda T, Nishida Y, Zhou Q, Yamamoto I, Kondo S, Azuma J. The impact of the CYP2D6 and CYP2C19 genotypes on venlafaxine pharmacokinetics in a japanese population. Eur J Clin Pharmacol. 2000;56(2):175–80.
Chen Y, Ke M, Xu J, Lin C. Simulation of the pharmacokinetics of oseltamivir and its active metabolite in Normal populations and patients with hepatic cirrhosis using physiologically based pharmacokinetic modeling. AAPS PharmSciTech. 2020;21(3):98.
Franek F, Jarlfors A, Larsen F, Holm P, Steffansen B. In vitro solubility, dissolution and permeability studies combined with semi-mechanistic modeling to investigate the intestinal absorption of desvenlafaxine from an immediate- and extended release formulation. Eur J Pharm Sci. 2015;77:303–13.
Lin HP, Sun D, Zhang X, Wen H. Physiologically based pharmacokinetic modeling for substitutability analysis of venlafaxine hydrochloride extended-release formulations using different release mechanisms: osmotic pump versus openable matrix. J Pharm Sci. 2016;105(10):3088–96.
Balhara A, Kale S, Singh S. Physiologically based pharmacokinetic (PBPK) modelling. In: Saharan VA, editor. Computer Aided Pharmaceutics and Drug Delivery: An Application Guide for Students and Researchers of Pharmaceutical Sciences. Singapore: Springer Nature Singapore; 2022. p. 255–84.
Tsamandouras N, Rostami-Hodjegan A, Aarons L. Combining the “bottom up” and “top down” approaches in pharmacokinetic modelling: fitting PBPK models to observed clinical data. Br J Clin Pharmacol. 2015;79(1):48–55.
Thelen K, Coboeken K, Willmann S, Burghaus R, Dressman JB, Lippert J. Evolution of a detailed physiological model to simulate the gastrointestinal transit and absorption process in humans, part 1: oral solutions. J Pharm Sci. 2011;100(12):5324–45.
Kawai R, Lemaire M, Steimer JL, Bruelisauer A, Niederberger W, Rowland M. Physiologically based pharmacokinetic study on a cyclosporin derivative, SDZ IMM 125. J Pharmacokinet Biopharm. 1994;22(5):327–65.
Willmann S, Lippert J, Schmitt W. From physicochemistry to absorption and distribution: predictive mechanistic modelling and computational tools. Expert Opin Drug Metab Toxicol. 2005;1(1):159–68.
Choi CI, Bae JW, Jang CG, Lee SY. Tamsulosin exposure is significantly increased by the CYP2D6*10/*10 genotype. J Clin Pharmacol. 2012;52(12):1934–8.
Byeon JY, Kim YH, Na HS, Jang JH, Kim SH, Lee YJ, et al. Effects of the CYP2D6*10 allele on the pharmacokinetics of atomoxetine and its metabolites. Arch Pharm Res. 2015;38(11):2083–91.
Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agúndez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther. 2020;107(1):154–70.
Botton MR, Whirl-Carrillo M, Del Tredici AL, Sangkuhl K, Cavallari LH, Agúndez JAG, et al. PharmVar GeneFocus: CYP2C19. Clin Pharmacol Ther. 2021;109(2):352–66.
Frye MA, Nemeroff CB. Pharmacogenomic testing for antidepressant treatment selection: lessons learned and roadmap forward. Neuropsychopharmacology. 2023;109:352–66.
Arnone D, Omar O, Arora T, Östlundh L, Ramaraj R, Javaid S, et al. Effectiveness of pharmacogenomic tests including CYP2D6 and CYP2C19 genomic variants for guiding the treatment of depressive disorders: systematic review and meta-analysis of randomised controlled trials. Neurosci Biobehav Rev. 2023;144:104965.
Vizirianakis IS, Mystridis GA, Avgoustakis K, Fatouros DG, Spanakis M. Enabling personalized cancer medicine decisions: the challenging pharmacological approach of PBPK models for nanomedicine and pharmacogenomics (review). Oncol Rep. 2016;35(4):1891–904.
PharmGKB. Annotation of DPWG Guideline for venlafaxine and CYP2D6 2019 [updated August 2019. Available from: https://www.pharmgkb.org/guidelineAnnotation/PA166104968.
Beunk L, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk AM, Guchelaar HJ, et al. Dutch pharmacogenetics working group (DPWG) guideline for the gene-drug interaction between CYP2D6, CYP3A4 and CYP1A2 and antipsychotics. Eur J Hum Genet. 2023. https://doi.org/10.1038/s41431-023-01347-3.
Gex-Fabry M, Balant-Gorgia AE, Balant LP, Rudaz S, Veuthey JL, Bertschy G. Time course of clinical response to venlafaxine: relevance of plasma level and chirality. Eur J Clin Pharmacol. 2004;59(12):883–91.
Lobello KW, Preskorn SH, Guico-Pabia CJ, Jiang Q, Paul J, Nichols AI, et al. Cytochrome P450 2D6 phenotype predicts antidepressant efficacy of venlafaxine: a secondary analysis of 4 studies in major depressive disorder. J Clin Psychiatry. 2010;71(11):1482–7.
Sakolsky DJ, Perel JM, Emslie GJ, Clarke GN, Wagner KD, Vitiello B, et al. Antidepressant exposure as a predictor of clinical outcomes in the treatment of resistant depression in adolescents (TORDIA) study. J Clin Psychopharmacol. 2011;31(1):92–7.
Karlsson L, Schmitt U, Josefsson M, Carlsson B, Ahlner J, Bengtsson F, et al. Blood-brain barrier penetration of the enantiomers of venlafaxine and its metabolites in mice lacking P-glycoprotein. Eur Neuropsychopharmacol. 2010;20(9):632–40.
Veefkind AH, Haffmans PM, Hoencamp E. Venlafaxine serum levels and CYP2D6 genotype. Ther Drug Monit. 2000;22(2):202–8.
Preskorn SH. Understanding outliers on the usual dose-response curve: venlafaxine as a way to phenotype patients in terms of their CYP 2D6 status and why it matters. J Psychiatr Pract. 2010;16(1):46–9.
Jiang F, Kim HD, Na HS, Lee SY, Seo DW, Choi JY, et al. The influences of CYP2D6 genotypes and drug interactions on the pharmacokinetics of venlafaxine: exploring predictive biomarkers for treatment outcomes. Psychopharmacology. 2015;232(11):1899–909.
Ahmed AT, Biernacka JM, Jenkins G, Rush AJ, Shinozaki G, Veldic M, et al. Pharmacokinetic-pharmacodynamic interaction associated with venlafaxine-XR remission in patients with major depressive disorder with history of citalopram / escitalopram treatment failure. J Affect Disord. 2019;246:62–8.
Acknowledgements
We express gratitude to all contributing authors for their participation and valuable discussions.
Funding
This work was supported by the Key Research and Development Program of Science and Technology Department of the Sichuan Province 111 Project (B18035).
Author information
Authors and Affiliations
Contributions
Ling Wang, Xuehua Jiang, Haitang Xie, and Chaozhuang Shen participated in the study's conceptualization. Chaozhuang Shen and Ling Wang drafted the manuscript. Hongyi Yang, Wenxin Shao, Liang Zheng, and Wei Zhang enriched the study through insightful discussions. All co-authors listed actively contributed to the modeling endeavors.
Corresponding author
Ethics declarations
Conflict of Interest
The authors affirm that they have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, C., Yang, H., Shao, W. et al. Physiologically Based Pharmacokinetic Modeling to Unravel the Drug-gene Interactions of Venlafaxine: Based on Activity Score-dependent Metabolism by CYP2D6 and CYP2C19 Polymorphisms. Pharm Res 41, 731–749 (2024). https://doi.org/10.1007/s11095-024-03680-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-024-03680-8