Abstract
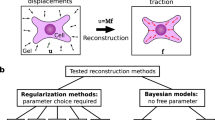
Traction force microscopy is a useful technique for measuring mechanical forces generated by cells. In this method, fluorescent nano beads are embedded in the elastic substrate of cell culture, on which cells are cultured. Then, cellular forces are estimated from bead displacements, which represent the force-induced deformation of the substrate under the cell. Estimating the forces from the bead displacements is not easy when the bead density is low or the locations of cellular attachments are unknown. In this study, we propose a Bayesian algorithm by introducing a prior force direction that is based on cellular morphology. We apply the Bayesian framework to synthetic datasets in conditions under which the bead density is low and cellular attachment points are unknown. We demonstrate that the Bayesian algorithm improves accuracy in force estimation compared with the previous algorithms.







Similar content being viewed by others
References
Kleeberger W, Bova GS, Nielsen ME, Herawi M, Chuang AY, Epstein JI, Berman DM (2007) Roles for the stem cell-associated intermediate filament nestin in prostate cancer migration and metastasis. Cancer Res 67(19):9199–9206
O’Hayre M, Salanga CL, Handel TM, Allen SJ (2008) Chemokines and cancer: migration, intracellular signalling and intercellular communication in the microenvironment. Biochem J 409:635–649
Calandra T, Bucala R (1997) Macrophage migration inhibitory factor (MIF): a glucocorticoid counter-regulator within the immune system. Crit Rev Immunol 17:77–88
Luster AD, Alon R, von Andrian UH (2005) Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol 6:1182–1190
Edmondson JC, Hatten ME (1987) Glial-guided granule neuron migration in vitro: a high-resolution time-lapse video microscopic study. J Neurosci 7(6):1928–1934
Tessier-Lavigne M, Goodman CS (1996) The molecular biology of axon guidance. Science 274:1123–1133
Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA (1998) Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA 95(16):9448–9453
Dickson BJ (2002) Molecular mechanisms of axon guidance. Science 298:1959–1964
Charron F, Tessier-Lavigne M (2005) Novel brain wiring functions for classical morphogens: a role as graded positional cues in axon guidance. Development 132:2251–2262
Beningo KA, Wang Y-L (2002) Flexible substrata for the detection of cellular traction forces. Trends Cell Biol 12:79–84
Roy P, Rajfur Z, Pomorski P, Jacobson K (2002) Microscope based-techniques to study cell adhesion and migration. Nat Cell Biol 4:E91–E96
Rieu JP, Barentin C, Maeda Y, Sawada Y (2005) Direct mechanical force measurements during the migration of dictyostelium slugs using flexible substrata. Biophys J 89:3563–3576
Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, Waterman CM (2008) Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J Cell Biol 183(6):999–1005
Stricker J, Sabass B, Schwarz US, Gardel ML (2010) Optimization of traction force microscopy for micron-sized focal adhesions. J Phys 22:194104
Bridgman PC, Dave S, Asnes CF, Tullio AN, Adelstein RS (2001) Myosin IIB is required for growth cone motility. J Neurosci 21(16):6159–6169
Suter DM, Forscher P (2001) Transmission of growth cone traction force through apCAM-cytoskeletal linkages is regulated by Src family tyrosine kinase activity. J Cell Biol 155(3):427–438
Chan CE, Odde DJ (2008) Traction dynamics of filopodia on compliant substrates. Science 322(5908):1687–1691
Toriyama M, Kozawa S, Sakumura Y, Inagaki N (2013) Conversion of a signal into forces for axon outgrowth through Pak1-mediated shootin1 phosphorylation. Curr Biol 23(6):529–534
Wang JHC, Lin JS (2007) Cell traction force and measurement methods. Biomech Model Mechanobiol 6:361–371
Landau LD, Lifshitz EM (1970) Theory of elasticity. Course of theoretical physics, 2nd edn. Pergamon, Oxford
Dembo M, Oliver T, Ishihara A, Jacobson K (1996) Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J 70:2008–2022
Dembo M, Wang YL (1999) Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J 76:2307–2316
Butler JP, Tolic-Norrelykkex IM, Fabry B, Fredberg JJ (2002) Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol 282:C595–C605
Yang Z, Lin JS, Chen J, Wang JH (2006) Determining substrate displacement and cell traction fields-a new approach. J Theor Biol 242:607–616
Schwarz US, Balaban NQ, Riveline D, Bershadsky A, Geiger B, Safran SA (2002) Calculation of forces at focal adhesions from elastic substrate data: the effect of localized force and the need for regularization. Biophys J 83:1380–1394
Sabass B, Gardel ML, Waterman CM, Schwarz US (2008) High resolution traction force microscopy based on experimental and computational advances. Biophys J 94:207–220
Forscher P, Smith SJ (1988) Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol 107:1505–1516
Lin CH, Espreafico EM, Mooseker MS, Forscher P (1996) Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron 16:769–782
Pollard TD, Borisy GG (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453–465
Pantaloni D, Clainche CL, Carlier MF (2001) Mechanism of actin-based motility. Science 292:1502–1506
Acknowledgments
This work was supported by a JSPS Grant-in-Aid for Scientific Research (C) (25330341) and Scientific Research on Innovative Areas (25102010).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kozawa, S., Sakumura, Y., Toriyama, M. et al. Bayesian Cell Force Estimation Considering Force Directions. Neural Process Lett 41, 191–200 (2015). https://doi.org/10.1007/s11063-013-9320-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11063-013-9320-y