Abstract
Purpose
Leptomeningeal carcinomatosis (LC) is a severe complication in the advanced stage of lung adenocarcinoma, with an extremely poor prognosis. Currently, the diagnosis of LC poses challenges. Serum exosomal miRNAs (microRNAs) have been demonstrated to possess potential as viable biomarkers. However, their value in the diagnosis of LC remains unclear.
Methods
In this study, serum samples were collected from lung adenocarcinoma patients with LC. The control groups consisted of patients with early-stage and advanced-stage lung adenocarcinoma without LC. Serum exosomes were isolated for high - throughput RNA sequencing to screen for differential miRNAs, and the results were validated in 123 samples. Subsequently, the receiver operating characteristic (ROC) curve was used to evaluate the diagnostic ability of exosomal miRNAs for LC.
Results
The results of miRNA sequencing revealed seven differentially enriched miRNAs (miRNA-1296-5p, miR-503-5p, miR-499a-5p, miR-374a-5p, miR-3173-5p, miR-370-3p and miR-885-3p). The ddPCR confirmed that the expression levels of miRNA-1296-5p, miR-499a-5p and miR-374a-5p were significantly elevated in LC, while miR-370-3p was significantly decreased (P < 0.05). ROC curve analysis showed that the AUC of the combination of miRNA-1296-5p, miR-499a-5p and miR-370-3p with CEA was 0.803 (P < 0.0001), displaying higher diagnostic power for LC.
Conclusion
This study suggests that the specific expression of miRNA-1296-5p, miR-499a-5p, miR-374a-5p and miR-370-3p in the serum exosomes of LC, which has diagnostic potential. And the combination of miRNA-1296-5p, miR-499a-5p and miR-370-3p with CEA can further enhance this potential.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Avoid common mistakes on your manuscript.
Introduction
leptomeningeal carcinomatosis (LC) is complication of advanced tumor, caused by cancer cells involvement of the leptomeninges [1]. Among solid tumors, patients with lung cancer, breast cancer, and melanoma are most commonly occur meningeal carcinomatosis [2]. About 3–8% of patients with non-small cell lung cancer (NSCLC) develop LC, and the incidence of LC appears to be increasing, possibly due to advances in diagnostic and treatment techniques that have led to more NSCLC patients achieving long-term survival, allowing LC to develop. The prognosis for LC is extremely poor. The median OS for untreated LC patients is only 4–6 weeks [3]. One reason for the dismal prognosis of LC could be the delay in treatment due to untimely diagnosis. Therefore, the diagnosis of LC without delay is considered crucial.
However, underdiagnosis and misdiagnosis of LC are common. LC diagnosis could be challenging. This is due to the frequently nonspecific symptoms of LC and the lack of sensitivity of currently diagnostic techniques [4]. There are two diagnostic methods used in clinical diagnosis of LC: magnetic resonance imaging (MRI) and cytological examination of cerebrospinal fluid (CSF) [5]. However, these two techniques are of limited sensitivity and often require repeated lumbar puncture and MRI. It is urgent to find new diagnostic methods to improve the detection rate of LC.
Exosomes are extracellular vesicles with a diameter of 40 to 160 nm, which have been proved to be useful as biomarkers for cancer early detection, early diagnosis, prognosis prediction [6]. Exosomes be used as tools for intercellular material and information exchange to transfer information molecules proteins, lipids, DNA, RNA and other molecular information between cells [7]. miRNAs are enriched in exosomes and play vital roles in gene regulation [8]. Exosomal miRNAs play key players in the occurrence and development of lung cancer and have been studied as diagnostic biomarkers for lung cancer metastasis [9, 10].
In this study, high-throughput sequencing was used to characterize the exosomal miRNA profiles of patients with LC and then their diagnostic potential was evaluated. Subsequently, the specifically enriched miRNAs were tested individually or in different combinations for their feasibility as diagnostic markers for LC.
Materials and methods
Clinical samples
All participants in this study were enrolled at the Second Hospital of Hebei Medical University from October 2018 to October 2023. Lung cancer patients with LC included in this study all had a positive diagnosis of CSF cytology. The control group consisted of individuals with early-stage lung cancer (stage I) and advanced-stage lung cancer (stage IV). The pathological type of the primary tumor in all patients was lung adenocarcinoma. This study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University, and written informed consent was obtained from each participant.
Isolation of exosomes
Exosomes from serum were isolated by size-exclusion chromatography. 1 ml of serum filtered through 0.8 μm was diluted, and then further purified using the Exosupur® chromatography column (Echobiotech, China). Subsequently, the sample was further eluted with 0.01 M PBS, and a total of 2 ml of eluate was collected according to the manufacturer’s instructions. The sample was concentrated to 200 µL by an Amicon® Ultra spin filter with a molecular weight of 100 kDa (Merck, Germany).
Identification of exosomes
Exosomes were observed by transmission electron microscopy (TEM). 10 µl exosomes was placed on a copper grid and left at room temperature for 5 min. After washing with PBS, it was fixed with 1% glutaraldehyde for 2 h. The exosomes were negatively stained with uranyl acetate solution for 5 min and then dried under an incandescent lamp for 5 min. Observation and photography were carried out under a transmission electron microscope (H-7650, Hitachi Ltd, Tokyo, Japan). The concentration and particle size of exosomes were measured using nanoparticle tracking analysis (NTA) technology. The ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany), equipped with a 405 nm laser, was used to detect vesicle suspensions with concentrations between 1 × 10⁷/ml and 1 × 10⁹/ml to determine the size and number of isolated particles. Exosomal proteins were detected by Western Blot (WB). The protein concentration was determined using a BCA protein assay kit (Thermo, MA, USA). The proteins were transferred to a PVDF membrane (Millipore, USA) and incubated with antibodies (TSG101, Abcam; ALIX, Abcam; CD63, Abcam; Calnexin, Abcam, GAPDH, CST). The protein signal was visualized using a chemiluminescence system (Bio-Rad, USA).
RNA library preparation and sequencing
Exosomal RNA was extracted using the miRNeasy® Mini Kit (QIAGEN, cat). The sequencing library was generated using the NEB Next Multiplex Small RNA Library Prep Set for Illumina (NEB, USA). Sequencing of the library was performed using the Illumina HiSeqTM2500 (Illumina, USA) instrument.
Evaluation of candidate exosomal miRNAs using DdPCR
The ddPCR reaction system was configured according to the instructions of the QX200 ddPCR EvaGreen Supermix. After preparing a water-in-oil emulsion, droplet PCR was performed. After PCR amplification, the QuantaSoft software was used to set up and analyze the sample information of the 96-well plate to complete the absolute quantification of the target RNA.
Statistical analysis
Statistical analysis and graphing of the relative expression levels of miRNA were performed using GraphPad Prism 9 software and SPSS 25.0 software. The comparison of means between two groups was conducted using the Student’s t test. A multivariable logistic regression analysis was employed to establish a miRNA combination model. The Receiver Operating Characteristic (ROC) curve was used to evaluate the diagnostic potential of exosomal miRNA for LC. The Youden index maximum was adopted to determine the sensitivity and specificity corresponding to the cut-off value of the ROC curve. <0.05 was considered statistically significant.
Results
Clinical characteristics of the participants
The experimental group included 42 lung adenocarcinoma patients with LC. The control group consisted of 41 stage - I and 40 stage - IV lung adenocarcinoma patients. There was no statistically significant difference in gender, age, and other general information between the experimental group and the control group. The clinical characteristics are shown in Table 1.
Characterization of exosomes isolated from serum of patients with LC
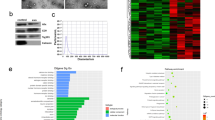
Exosomes were extracted from the serum of patients with LC using the size-exclusion chromatography (SEC). Observation by TEM revealed micromembrane‑coated vesicular structures with a single distribution, and had a bilayer membrane structure (Fig. 1A). The particle size distribution and concentration of exosome samples were measured by NTA. The results showed that 95% of the exosome particles were distributed around 150 nm (Fig. 1B). Exosomal marker proteins were detected by WB, revealing that the common marker proteins of exosomes, namely Alix, TSG101 and CD63, were present on the exosome membrane, while the exosome negative protein Calnexin was not detected (Fig. 1C) (The figures in this part are from our previously published article in Molecular Cancer Research). These results suggested that the extracted vesicles were exosomes.
Identification of serum exosomes and sequencing heatmap. (A) Transmission electron microscopy revealed the morphology and size of exosomes. Scale bar, 200 nm. (B) Nanoparticle Tracking Analysis of the Size Distribution and Concentration of Exosomes. (C) Western blot was used to detect specific exosomal markers of exosomes, including ALIX, TSG101, and CD63, as well as the negative protein Calnexin. (D) The Volcano Plot of 7 DEGs between control and LC. (E) Expression levels of 7 DEGs were analyzed in the heatmap of 9 samples
High-throughput sequencing was used to screen serum Exosomal miRNAs
Nine serum samples (including three cases each of patients with LC, early-stage lung cancer and advanced-stage lung cancer) were collected. Then exosomes were isolated and total RNA was extracted for high-throughput small RNA (sRNA) sequencing. In terms of the identification of known miRNAs, the clean read sequences obtained by sequencing were aligned with the mature miRNA sequences in the known miRNA database miRBase (v22). The miRDeep2 software package was utilized to predict new miRNAs. Through analysis, a total of 913 miRNAs were obtained from all samples, among which 892 were known miRNAs and 21 were newly predicted miRNAs. The edgeR software was used for the differential miRNA expression analysis. The analysis criteria were set as|log2 (fold change, FC)| ≥ 0.58 and P ≤ 0.05. Through this method, miRNAs with differentially enriched amog different groups were identified. As a result, 7 miRNAs with significant differences in expression among the groups were screened out. Among them, 4 miRNAs (miRNA-1296-5p, miR-503-5p, miR-499a-5p and miR-374a-5p) were enriched in high abundance, and 3 miRNAs (miR-3173-5p, miR-370-3p and miR-885-3p) were down-regulated (Fig. 1D and E). KEGG analyses demonstrated the involvement of differentially enriched miRNAs in pathways in cancer, antigen processing and presentation, miRNAs in cancer, primary immunodeficiency and notch signaling pathway (Fig. 2A). The results of GO analysis showed that miRNAs had a strong correlation with TAP1 binding, TAP2 binding, MHC class I protein binding and peptide antigen binding (Fig. 2B). The analysis of biological processes mainly focuses on the defense response and antigen processing and presentation of endogenous peptide antigen via MHC class I (Fig. 2C).
Analysis of miRNA expression differences and verification by droplet digital PCR (ddPCR)
DdPCR was used to detect the expression levels of the above seven candidate miRNAs (miRNA-1296-5p, miR-503-5p, miR-499a-5p, miR-374a-5p, miR-3173-5p, miR-370-3p and miR-885-3p) in 123 serum exosome samples (including 42 cases of LC, 41 cases of early lung cancer and 40 cases of advanced lung cancer). The difference rate of miRNA enrichment levels in the LC compared with those in the control were calculated. It was found that the enrichment levels of miRNA-1296-5p, miR-499a − 5p and miR-374a-5p were significantly up-regulated in the serum exosomes of LC patients (Fig. 3B and E), while the enrichment level of miR-370-3p was significantly down-regulated (P < 0.05) (Fig. 3A). There were no differences in the enrichment of miR-503-5p, miR-885-3p and miR-3173-5p (Supplementary Fig. 1A-1 C). These results suggest that miRNA-1296-5p, miR-499a-5p, miR-374a-5p and miR-370-3p may be useful markers for diagnosis of LC.
ROC curve analysis of the diagnostic potential of miRNA for LC
ROC curves were constructed to evaluate the diagnostic potential of the differentially enriched miRNAs in serum exosomes for LC. Meanwhile, the 95% confidence intervals (CIs), the area under the curve (AUC) values, as well as the sensitivity and specificity were calculated. The results demonstrated that miR-374a-5p yielded an AUC of 0.63 (95% CI, 0.505–0.755) (Fig. 4A). MiR-370-3p yielded an AUC of 0.67 (95% CI, 0.5526–0.78) (Fig. 4B). The AUC of miR-499a-5p was 0.698 (95% CI, 0.584–0.812) (Fig. 4C). MiRNA-1296-5p displayed an AUC of 0.712 (95% CI, 0.606–0.823) (Fig. 4D).
ROC curve analysis of the diagnostic potential of miRNA combinations for LC
Regression analysis models of multiple miRNA combinations were constructed to improve the diagnostic potential for LC. The diagnostic potential of these combinations was analyzed using the ROC curve. The results demonstrated that the diagnostic performance of multiple miRNA combinations for LC was superior to that of a single miRNA (Supplementary Fig. 2A-2I). In particular, the combination of miRNA-1296-5p, miR-499a-5p and miR-370-3p had an AUC of 0.778 (95% CI, 0.676–0.876) for the diagnosis of LC (Fig. 5A). Furthermore, the introduction of carcinoembryonic antigen (CEA) into the miRNAs effectively improved the diagnostic rate of LC (Fig. 5B and D). Specifically, the AUC for the combined diagnosis of LC by CEA and miRNA-1296-5p was 0.746 (95% CI, 0.634–0.858) (Fig. 5E). The diagnostic efficiency was significantly improved when the miRNA combination was combined with CEA (Supplementary Fig. 3A-3I). MiRNA-1296-5p, miR-499a-5p and miR-370-3p were combined with CEA, the resulting AUC was 0.803 (95% CI, 0.708–0.898) (Fig. 5F), which had a more excellent diagnostic effect on LC.
ROC curves of exosomal miRNAs combinations for the diagnosis of LC. ROC curve analysis of (A) miRNA-1296-5p and miR-499a-5p and miR-370-3p, (B) miR-374a-5p combined with CEA, (C) miR-499a-5p combined with CEA, (D) miR-370-3p combined with CEA, (E) miRNA-1296-5p combined with CEA, (F) miRNA-1296-5p and miR-499a-5p and miR-370-3p combined with CEA
Discussion
The prognosis of LC has an unusually ominous. Early and accurate diagnosis of LC is important, but the current clinical diagnostic methods remain inadequate [11]. In this study, a comprehensive analysis of the miRNA profiles of serum exosomes was conducted by using miRNA-seq technology, and seven differentially enriched exosomal miRNAs were identified. Subsequently, further verification was carried out in the serum exosomes of 123 cases. Eventually, it was found that the expressions of miRNA-1296-5p, miR-499a-5p and miR-374a-5p in the serum exosomes of LC patients were significantly increased, while the expression of miR-370-3p was significantly decreased. Through ROC curve analysis, it was discovered that all four miRNAs demonstrated the diagnostic potential for LC. And the optimal combination was determined to be miRNA-1296-5p, miR-499a-5p and miR-370-3p combined with CEA for diagnosing LC.
Currently, neurological MRI and CSF cytology are the most frequently utilized diagnostic methods for LC. Positive CSF cytology is considered to be the gold standard for the diagnosis of LC. Nevertheless, these two methods are short of specificity and sensitivity [12]. A growing number of studies are seeking ways to diagnose LC using liquid biopsies, most of which are based primarily on CSF of patients. Especially the detection of cytometry circulating tumor cells (CTC) in CSF has aroused great interest among people. In a relevant study, researchers carried out a comparative analysis on the performance of CTC detection and CSF cytology. The results of the study demonstrated that CTC detection showed relatively high detection efficiency. Its sensitivity reached 94% (95% CI 80–99), and its specificity was as high as 100% (with the 95% CI 91–100). In contrast, the sensitivity of cytological examination was 76% (95% CI 58–89), which was slightly inferior to that of CTC detection in terms of overall detection performance [13]. In addition, CSF circulating tumor DNA (ct-DNA) has been proved to effectively increase the sensitivity and specificity for diagnosing LC. One study found through ultra-low-pass whole-genome sequencing (ulpWGS) that the CSF ctDNA fraction in patients with breast cancer leptomeningeal metastasis (BC-LC) was significantly higher than that in patients without metastasis, and ctDNA (fraction ≥ 0.10) could be detected in all BC-LC positive patients, demonstrating the potential for timely and accurate diagnosis of LC [14]. Another experiment discovered that there were differences between the results of CSF ctDNA analysis and CSF cytology among breast cancer patients with suspected LC, suggesting that CSF ctDNA can supplement the diagnosis of LC [15]. Single-cell RNA sequencing (scRNA-seq) and cell-free RNA (cfRNA) technologies have also been used to diagnose LC. It has been found that MUC1 and CEACAM6 are highly enriched in LC cells, and CEACAM6 can be detected in the cfRNA of patients’ CSF, with a detection sensitivity of 88.24% and a specificity of 100% [16].
This study selected exosomes for the research on the diagnostic method of LC based on several advantages of exosomes: they are widely sourced and easily to obtain, contain abundant information molecules, possess high stability, and can freely cross the blood-brain barrier [17]. Multiple studies have shown that the differential expression of exosome contents is closely related to cancer metastasis and are promising biomarkers for diagnosis/prognosis. Exosomal integrins have been found to predict organ-specific metastasis. Integrins α6β4 and α6β1 are related to lung metastasis, and integrin αvβ5 is associated with liver metastasis [18]. Studies have found that high levels of exosomal Tim-3 and galectin 9 in plasma exosomes are positively correlated with tumor lymph node metastasis and distant metastasis [19]. Exosomal MUC5B and SELL can serve as biomarkers for diagnosing brain metastasis of lung cancer, while APOH may be a potential diagnostic biomarker for liver metastasis of lung cancer [20]. Exosomal miR-151a-3p and miR-877-5p may also be biomarkers for predicting bone metastasis of lung cancer [21]. The exosome miRNAs associated with LC were paid more attention. RNA expression profiles obtained in 472 human CSF exosomes showed that the exosome miR-21 could be used as a biomarker for lung cancer LC [22]. Li’s experiment found that three exosome miRNAs with high expression from CSF (miR-183-5p, miR-96-5p and miR-182-5p) could diagnose LC [23]. There are few experiments based on blood exosomes. Xu found miR-483-5p in serum exosomes of 7 patients with LC, and miR-342-5p may participate in LC of lung cancer and may replace CSF in predicting NSCLC-LC, however, their samples were insufficient [24].
Our research findings indicate that serum exosomal miR-1296–5p, miR-499-5p, miR-374a-5p and miR-370-3p are differentially enriched in lung adenocarcinoma patients with LC. MiR-374a-5p acts as an oncogenic factor and plays a role in the process of cancer [25]. Our previous research explored that exosomal miR-374a-5p derived from NSCLC in serum participates in the formation of the pre-metastatic niche and promotes LC by targeting the expression of ADD3 in brain microvascular endothelial cells, altering the distribution of tight junction proteins, and regulating the permeability of the blood-brain barrier [26]. MiRNA-1296-5p, miRNA-499a-5p and MiR-370-3p have both tumor-inducing and tumor-suppressing effects in different cancers [27,28,29]. It has been reported that miRNA-1296-5p is related to processes such as cell adhesion molecules (CAMs) and transmigration across endothelial cells [30]. One study showed that the expression of miR-499a-5p was significantly upregulated in patients with vascular endothelial dysfunction [31]. MR-370-3p is closely related to endothelial dysfunction and participates in the regulation of blood-brain barrier (BBB) permeability [32]. It has been confirmed that miRNA are involved in the regulation of vascular endothelial function [33, 34]. Previous studies have shown that these three miRNAs all seem to be associated with endothelial dysfunction, which may be related to the mechanism of action during the process of liver cirrhosis (LC). We will explore this in future research.
Conclusion
This study found that miRNA-1296-5p, miR-499a-5p, and miR-374a-5p are highly enriched in LC serum exosomes, while miR-370-3p is lowly enriched. The combination of miRNA-1296-5p, miR-499a-5p, miR-370-3p, and CEA shows relatively high diagnostic sensitivity for LC diagnosis. Future research on the molecular mechanisms of miRNA-1296-5p, miR-499a-5p, and miR-370-3p in LC will further improve the understanding of their functional roles. This study has found the potential role of these miRNAs in disease prediction, however, due to the limited clinical samples and individual differences, it is necessary to increase the sample size to further validate their diagnostic sensitivity and specificity.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- LC:
-
Leptomeningeal carcinomatosis.
- NSCLC:
-
Non-small cell lung cancer.
- CSF:
-
Cerebrospinal fluid.
- TEM:
-
Transmission electron microscopy.
- NTA:
-
Nanoparticle tracking analysis.
- WB:
-
Western blot.
- SEC:
-
Size-exclusion chromatography.
- CIs:
-
Confidence intervals.
- AUC:
-
Area under the curve.
- ROC:
-
Receiver operating characteristic curve.
- CTC:
-
Cytometry circulating tumor cells.
- ct-DNA:
-
Circulating tumor DNA.
- BBB:
-
Blood-brain barrier.
References
Jan R, Adrienne B (2024) The path to leptomeningeal metastasis. Nat Rev Cancer 24(7). https://doi.org/10.1038/s41568-024-00700-y
Ahmad O, Hannah W, Debarati B, Nicholas M, Michael G, Stuart AG et al (2024) Leptomeningeal metastatic disease: new frontiers and future directions. Nat Rev Clin Oncol 22(2). https://doi.org/10.1038/s41571-024-00970-3
Sehhoon P, Richard B, Hyun Ae J, Jong-Mu S, Se-Hoon L, Jin Seok A et al (2024) Phase II Efficacy and Safety of 80 mg Osimertinib in Patients With Leptomeningeal Metastases Associated With Epidermal Growth Factor Receptor Mutation-Positive Non-Small Cell Lung Cancer (BLOSSOM). J Clin Oncol 42(23). https://doi.org/10.1200/jco.24.00708
Lindsay A, John WMM, Martin J, vdB, Peter AESS, Stefan S, Agnes J (2018) Novel methods to diagnose leptomeningeal metastases in breast cancer. Neuro Oncol 21(4). https://doi.org/10.1093/neuonc/noy186
Haiying C, Roman P-S (2018) Leptomeningeal metastases in non-small-cell lung cancer. Lancet Oncol 19(1). (https://doi.org/10.1016/s1470-2045(17)30689-7)
Raghu K, Kathleen MM (2023) The role of extracellular vesicles in cancer. Cell 186(8). https://doi.org/10.1016/j.cell.2023.03.010
Raghu K, Valerie SL (2020) The biology, function, and biomedical applications of exosomes. Science 367(6478). https://doi.org/10.1126/science.aau6977
Sushmaa Chandralekha S, K Auxzilia P, Durairaj S (2023) MicroRNAs as important players in regulating cancer through PTEN/PI3K/AKT signalling pathways. Biochim Biophys Acta Rev Cancer 1878(3). https://doi.org/10.1016/j.bbcan.2023.188904
Wei Y, Xiwei W, Weiying Z, Miranda YF, Minghui C, Juan L et al (2018) Cancer-cell-secreted Exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol 20(5). https://doi.org/10.1038/s41556-018-0083-6
Mingyang J, Yiji J, Kaicheng L, Fu G, Ke Z, Mingjing X et al (2023) Exosome-mediated miR-144-3p promotes ferroptosis to inhibit osteosarcoma proliferation, migration, and invasion through regulating ZEB1. Mol Cancer 22(1). https://doi.org/10.1186/s12943-023-01804-z
E LR MW, vdB M (2023) Leptomeningeal metastasis from solid tumours: EANO-ESMO clinical practice guideline for diagnosis, treatment and follow-up. ESMO Open 8(5). https://doi.org/10.1016/j.esmoop.2023.101624
Jyoti M, Isa M, Gregory G, Jeremy F, Arin N, Natalie G et al (2024) Targeting CNS metastases in Non-Small cell lung Cancer with evolving approaches using molecular markers: A review. JAMA Oncol 11(1). https://doi.org/10.1001/jamaoncol.2024.5218
Mark TJ, vB, Dick P, Bojana MK, Mijke B, Karolina S, Dorothé TCL et al (2020) Circulating epithelial tumor cell analysis in CSF in patients with leptomeningeal metastases. Neurology 94(5). https://doi.org/10.1212/wnl.0000000000008751
Amanda F, Marjan I, Adam M, Lucy C, Thanussuyah A, Angela C et al (2021) Assessing CSF ctdna to improve diagnostic accuracy and therapeutic monitoring in breast Cancer leptomeningeal metastasis. Clin Cancer Res 28(6). https://doi.org/10.1158/1078-0432.Ccr-21-3017
Leticia DM-A, Regina M, Charlotte KYN, Britta W, Francisco M-R, Davis T et al (2015) Cerebrospinal fluid-derived Circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 6(0). https://doi.org/10.1038/ncomms9839
Yingmei L, Dina P, Layton L, Ian David C, Eli J, Lina Khav K et al (2021) Comprehensive RNA analysis of CSF reveals a role for CEACAM6 in lung cancer leptomeningeal metastases. NPJ Precis Oncol 5(1). https://doi.org/10.1038/s41698-021-00228-6
Pegtel D, Gould S, Exosomes (2019) Annu Rev Biochem 88:487–514. https://doi.org/10.1146/annurev-biochem-013118-111902
Hoshino A, Costa-Silva B, Shen T, Rodrigues G, Hashimoto A, Tesic Mark M et al (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527(7578):329–335. https://doi.org/10.1038/nature15756
Liu Q, Xiang Y, Yuan S, Xie W, Li C, Hu Z et al (2018) Plasma exosome levels in non-small-cell lung cancer: correlation with clinicopathological features and prognostic implications. Cancer biomarkers: section A of disease markers. 22(2):267–274. https://doi.org/10.3233/cbm-170955
Li S, Qu Y, Liu L, Zhang X, He Y, Wang C et al (2023) Comparative proteomic profiling of plasma exosomes in lung cancer cases of liver and brain metastasis. Cell Bioscience 13(1):180. https://doi.org/10.1186/s13578-023-01112-5
Kun Z, Changji J, Jin W, Weiye S, Xiaoying W, Yan S et al (2024) Exosomal hsa-miR-151a-3p and hsa-miR-877-5p are potential novel biomarkers for predicting bone metastasis in lung cancer. Aging 15(24). https://doi.org/10.18632/aging.205314
Lee K, Im J, Lin W, Gwak H, Kim J, Yoo B et al (2020) Nanoparticles in 472 human cerebrospinal fluid: changes in extracellular vesicle concentration and miR-21 expression as a biomarker for leptomeningeal metastasis. Cancers 12(10). https://doi.org/10.3390/cancers12102745
Li H, Xia M, Zheng S, Lin Y, Yu T, Xie Y et al (2024) Cerebrospinal fluid Exosomal MicroRNAs as biomarkers for diagnosing or monitoring the progression of non-small cell lung cancer with leptomeningeal metastases. Biotechnol Genet Eng Rev 40(1):359–380. https://doi.org/10.1080/02648725.2023.2183613
Xu Q, Ye L, Huang L, Zhou L, Chen X, Ye M et al (2021) Serum Exosomal MiRNA might be a novel liquid biopsy to identify leptomeningeal metastasis in Non-Small cell lung Cancer. OncoTargets Therapy 14:2327–2335. https://doi.org/10.2147/ott.S291611
Ji R, Zhang X, Gu H, Ma J, Wen X, Zhou J et al (2019) miR-374a-5p: A new target for diagnosis and drug resistance therapy in gastric Cancer. Mol Therapy Nucleic Acids 18:320–331. https://doi.org/10.1016/j.omtn.2019.07.025
Jie J, Yumeng C, Huicong N, Yanli L, Xiaojie W, Xuejiao Q et al (2024) NSCLC extracellular vesicles containing miR-374a-5p promote leptomeningeal metastasis by influencing Blood–Brain barrier permeability. Mol Cancer Res 22(8). https://doi.org/10.1158/1541-7786.Mcr-24-0052
Qing C, Lin Z, De M, Juan H, Yuxin L, Jie W et al (2022) LncRNA GAS6-AS1 facilitates tumorigenesis and metastasis of colorectal cancer by regulating TRIM14 through miR-370-3p/miR-1296-5p and FUS. J Transl Med 20(1). https://doi.org/10.1186/s12967-022-03550-0
Shan H, Ziming L, Yongfeng Y, Qingyu Z, Yirui C, Wenxiang J et al (2019) Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp Cell Res 379(2). https://doi.org/10.1016/j.yexcr.2019.03.035
Siqi P, Yutong C, Ting L, Junjie M, Pengfei Y, Baojia Z et al (2022) Hsa-microRNA-370-3p targeting snail and Twist1 suppresses IL-8/STAT3-driven hepatocellular carcinoma metastasis. Cancer Sci 113(12). https://doi.org/10.1111/cas.15571
Mengying N, Hong L, Xu L, Xiaoqian Y, Aijun M, Xudong P et al (2021) Circulating Exosomal MiRNAs as novel biomarkers perform superior diagnostic efficiency compared with plasma MiRNAs for Large-Artery atherosclerosis stroke. Front Pharmacol 12(0). https://doi.org/10.3389/fphar.2021.791644
Ilona H, Katerina K, Lenka D, Ladislav K (2020) Evaluation of vascular endothelial function in young and Middle-Aged women with respect to a history of pregnancy, Pregnancy-Related complications, classical cardiovascular risk factors, and epigenetics. Int J Mol Sci 21(2). https://doi.org/10.3390/ijms21020430
Caifeng G, Weichun M, Kunlun W, Mingqiang G, Junfeng C, Feng Z et al (2023) Exosomal miR-370-3p increases the permeability of blood-brain barrier in ischemia/reperfusion stroke of brain by targeting MPK1. Aging 15(6). https://doi.org/10.18632/aging.204573
Sushmaa Chandralekha S, Auxzilia PK, Durairaj S (2024) MicroRNA-510-3p regulated vascular dysfunction in preeclampsia by targeting vascular endothelial growth factor A (VEGFA) and its signaling axis. Placenta 153(0). https://doi.org/10.1016/j.placenta.2024.05.135
Mani P, R M V MB, Durairaj PSSRS (2018) Dissecting the role of miR-21 in different types of stroke. Gene 681(0). https://doi.org/10.1016/j.gene.2018.09.048)
Acknowledgements
The authors thank Beijing Enze Kangtai Biotechnology Co., Ltd (China) for its support of RNA-seq.
Funding
This study was supported by Medical science research project of Hebei Provincial Health Commission of China (No. 20251629).
Author information
Authors and Affiliations
Contributions
Junjuan Qin and Jiasi Zhang collected the clinical sample. Jie Jin detected the miRNAs in serum exosomes, Junjuan Qin, Xuejiao Qi, Jiasi Zhang, YingLu Zhang collected the data and conducted the statistical analyses. Jie Jin wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics statement
The research using human serum samples was approved by the Ethics Committee of the Second Hospital of Hebei Medical University and was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.



Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jin, J., Qin, J., Qi, X. et al. Serum exosomal miRNA contributes to the diagnosis of leptomeningeal carcinomatosis. J Neurooncol (2025). https://doi.org/10.1007/s11060-025-04999-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11060-025-04999-x