Abstract
Background
Candida auris is a multidrug-resistant pathogen that causes nosocomial outbreaks and high mortality. We conducted this study to investigate the molecular mechanisms of antifungal resistance in our clinical isolate of C. auris with a high level of resistance to three main classes of antifungals.
Material and Methods
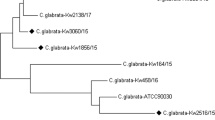
A clinical C. auris isolate was identified by MALDI-TOF MS and antifungal susceptibilities were determined by the Sensititre YeastOne YO10 panel. After sequencing the whole genome of the microorganism with Oxford Nanopore NGS Technologies, a phylogenetic tree was drawn as a cladogram to detect where the C. auris clade to this study’s assembly belongs.
Results
The C. auris isolate in this study (MaCa01) was determined to be a part of the clade I (South Asian). The resistance-related genes indicated that MaCa01 would most likely be highly resistant to fluconazole (CDR1, TAC1b, and ERG11), none or little resistant to amphotericin B (AmpB) and echinocandins, and sensitive to flucytosine. The mutations found in the above-mentioned genes in the Türkiye C. auris isolate reveals an antifungal resistance pattern. This molecular resistance pattern was found consistent with the interpretation of MIC values of the antifungals according to CDC tentative breakpoints.
Conclusion
We detected the well-known antifungal resistance mutations, responsible for azole resistance in C. auris. Despite no ERG2, ERG6, and FKS mutation identified, the isolate was found to be resistant to AmpB and caspofungin based on the CDC tentative breakpoints which could be related to unidentified mutations.

Similar content being viewed by others
References
Di Pilato V, Codda G, Ball L, Giacobbe DR, Willison E, Mikulska M, et al. Molecular epidemiological investigation of a nosocomial cluster of C. auris: evidence of recent emergence in Italy and ease of transmission during the COVID-19 pandemic. J Fungi (Basel). 2021;7(2):140. https://doi.org/10.3390/jof7020140.
Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019;25(9):1780–1. https://doi.org/10.3201/eid2509.190686.
Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64(2):134–40. https://doi.org/10.1093/cid/ciw691.
Centers for Disease Control and Prevention (CDC): C. auris: antifungal susceptibility testing and interpretation. https://www.cdc.gov/fungal/candida-auris/c-auris-antifungal.html (2020). Accessed 29 May 2020.
Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, et al. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018;73(4):891–9. https://doi.org/10.1093/jac/dkx480.
Magobo RE, Corcoran C, Seetharam S, Govender NP. Candida auris-associated candidemia, South Africa. Emerg Infect Dis. 2014;20(7):1250–1. https://doi.org/10.3201/eid2007.131765.
Chow NA, Munoz JF, Gade L, Berkow EL, Li X, Welsh RM, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio. 2020. https://doi.org/10.1128/mBio.03364-19.
Chaabane F, Graf A, Jequier L, Coste AT. Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol. 2019;10:2788. https://doi.org/10.3389/fmicb.2019.02788.
Rybak JM, Barker KS, Munoz JF, Parker JE, Ahmad S, Mokaddas E, et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect. 2022;28(6):838–43. https://doi.org/10.1016/j.cmi.2021.11.024.
Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7(1):43. https://doi.org/10.1038/s41426-018-0045-x.
Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–6. https://doi.org/10.1038/s41587-019-0072-8.
Gosselin SFM, Feng Y, Gogarten JP. Expanding the utility of sequence comparisons using data from whole genomes. bioRxiv. 2020. https://doi.org/10.1101/2020.01.15.908137.
Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. https://doi.org/10.1093/nar/gkab301.
Nash A, Sewell T, Farrer RA, Abdolrasouli A, Shelton JMG, Fisher MC, et al. MARDy: mycology antifungal resistance database. Bioinformatics. 2018;34(18):3233–4. https://doi.org/10.1093/bioinformatics/bty321.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. https://doi.org/10.1093/nar/gky427.
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. https://doi.org/10.1002/pro.3943.
Rhodes J, Fisher MC. Global epidemiology of emerging Candida auris. Curr Opin Microbiol. 2019;52:84–9. https://doi.org/10.1016/j.mib.2019.05.008.
Frias-De-Leon MG, Hernandez-Castro R, Vite-Garin T, Arenas R, Bonifaz A, Castanon-Olivares L, et al. Antifungal resistance in Candida auris: molecular determinants. Antibiotics (Basel). 2020;9(9):568. https://doi.org/10.3390/antibiotics9090568.
Rybak JM, Munoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, et al. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. MBio. 2020. https://doi.org/10.1128/mBio.00365-20.
Carolus H, Pierson S, Munoz JF, Subotic A, Cruz RB, Cuomo CA, et al. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. MBio. 2021. https://doi.org/10.1128/mBio.03333-20.
Rybak JM, Doorley LA, Nishimoto AT, Barker KS, Palmer GE, Rogers PD. Abrogation of triazole resistance upon deletion of CDR1 in a clinical isolate of Candida auris. Antimicrob Agents Chemother. 2019. https://doi.org/10.1128/AAC.00057-19.
Centers for Disease Control and Prevention (CDC): Antibiotic resistance threats in the United States. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (2019). Accessed 26 March 2019.
Kean R, Delaney C, Sherry L, Borman A, Johnson EM, Richardson MD, et al. Transcriptome assembly and profiling of Candida auris reveals novel insights into biofilm-mediated resistance. mSphere. 2018. https://doi.org/10.1128/mSphere.00334-18.
Lockhart SR. Candida auris and multidrug resistance: defining the new normal. Fungal Genet Biol. 2019;131:103243. https://doi.org/10.1016/j.fgb.2019.103243.
Arendrup MC, Patterson TF. Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J Infect Dis. 2017;216(suppl_3):S445–51. https://doi.org/10.1093/infdis/jix131.
Asadzadeh M, Mokaddas E, Ahmad S, Abdullah AA, de Groot T, Meis JF, et al. Molecular characterisation of Candida auris isolates from immunocompromised patients in a tertiary-care hospital in Kuwait reveals a novel mutation in FKS1 conferring reduced susceptibility to echinocandins. Mycoses. 2022;65(3):331–43. https://doi.org/10.1111/myc.13419.
Beyda ND, Lewis RE, Garey KW. Echinocandin resistance in Candida species: mechanisms of reduced susceptibility and therapeutic approaches. Ann Pharmacother. 2012;46(7–8):1086–96. https://doi.org/10.1345/aph.1R020.
Walker LA, Gow NA, Munro CA. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob Agents Chemother. 2013;57(1):146–54. https://doi.org/10.1128/AAC.01486-12.
Fayed B, Jayakumar MN, Soliman SSM. Caspofungin-resistance in Candida auris is cell wall-dependent phenotype and potential prevention by zinc oxide nanoparticles. Med Mycol. 2021;59(12):1243–56. https://doi.org/10.1093/mmy/myab059.
Funding
This study was supported by Gilead Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared no potential conflicts of interest concerning this article's authorship and/or publication.
Ethical Approval
Marmara University School of Medicine Institutional Ethical Review Board issued approval 09.2021.578. The collected data did not include any patient or employee identifying information.
Additional information
Handling Editor: Ferry Hagen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Erturk Sengel, B., Ekren, B.Y., Sayin, E. et al. Identification of Molecular and Genetic Resistance Mechanisms in a Candida auris Isolate in a Tertiary Care Center in Türkiye. Mycopathologia 188, 929–936 (2023). https://doi.org/10.1007/s11046-023-00787-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11046-023-00787-1