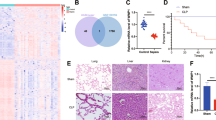
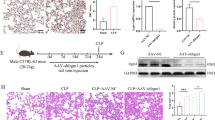
Abstract
Introduction
The purpose of this investigation was to elucidate the functions of TRIM11 and HOXB9 in the pathogenesis of sepsis, focusing on their influence on inflammation, apoptosis, and the NF-κB signaling pathway.
Material and methods
Through public databases, TRIM family genes related to sepsis were screened, and TRIM11 was evaluated as a sepsis biomarker through ROC analysis. The UbiBrowser database screened TRIM11 downstream genes and identified HOXB9 as an essential target. THP-1 cells were stimulated by Lipopolysaccharide (LPS) to induce inflammation and simulate sepsis. Flow cytometry, Enzyme-linked immunosorbent assay, and Western blot experiments were used to detect changes in cell apoptosis rate, apoptosis-related proteins, and inflammatory cytokines after TRIM11 and HOXB9 were silenced. Additionally, we investigated the ubiquitination interaction between TRIM11 and HOXB9 and their effects on the NF-κB signaling pathway.
Results
Our findings demonstrated that sepsis patient samples had elevated levels of TRIM11 expression and had high clinical diagnostic value. Functional experiments showed that the knockdown of TRIM11 significantly alleviated LPS-induced THP-1 cell apoptosis and inflammation, while the knockdown of HOXB9 did the opposite. The simultaneous downregulation of TRIM11 and HOXB9 balanced these responses, suggesting they play a key role in regulating sepsis-associated inflammation and apoptosis. In addition, TRIM11 regulated the NF-κB signaling pathway by reversing HOXB9-induced activation through ubiquitination, suggesting a novel regulatory mechanism in the pathogenesis of sepsis.
Conclusions
Our findings highlight the interaction between TRIM11 and HOXB9 in regulating inflammation and apoptosis pathways, providing new insights into sepsis treatment.








Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Cecconi M, Evans L, Levy M, Rhodes A (2018) Sepsis and septic shock. Lancet 392(10141):75–87
Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR et al (2020) Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 395(10219):200–211
Minasyan H (2019) Sepsis: mechanisms of bacterial injury to the patient. Scandinavian J Trauma Resusc Emerg Med 27(1):19
Williams B, Zou L, Pittet JF, Chao W (2024) Sepsis-induced coagulopathy: a comprehensive narrative review of pathophysiology, clinical presentation, diagnosis, and management strategies. Anesth Analg 138(4):696–711
Salomão R, Ferreira B, Salomão M, Santos S, Azevedo L, Brunialti M (2019) Sepsis: evolving concepts and challenges. Braz J Med Biol Res 52:e8595
Ranjit S, Kissoon N (2021) Challenges and solutions in translating sepsis guidelines into practice in resource-limited settings. Transl Pediatr 10(10):2646
van der Slikke EC, Beumeler LF, Holmqvist M, Linder A, Mankowski RT, Bouma HR (2023) Understanding post-sepsis syndrome: how can clinicians help? Infect Drug Resistance 16:6493–6511
Aizaz M, Kiani YS, Nisar M, Shan S, Paracha RZ, Yang G (2023) Genomic analysis, evolution and characterization of E3 ubiquitin protein ligase (TRIM) gene family in common carp (Cyprinus carpio). Genes 14(3):667
Wang L, Ning S (2021) TRIMming type I interferon-mediated innate immune response in antiviral and antitumor defense. Viruses 13(2):279
Zhou G, Wu W, Yu L, Yu T, Yang W, Wang P et al (2018) Tripartite motif-containing (TRIM) 21 negatively regulates intestinal mucosal inflammation through inhibiting TH1/TH17 cell differentiation in patients with inflammatory bowel diseases. J Allerg Clin Immunol. 142(4):1218-1228.e12
Zhang J, Zhu J, Chen X, Xia H, Yang L (2022) E3 ubiquitin ligase Trim33 ubiquitylates Annexin A2 to promote NF-κB induced skin inflammation in psoriasis. J Dermatol Sci 107(3):160–168
Xie X, Wang F, Li X (2022) Inhibition of TRIM14 protects cerebral ischemia/reperfusion injury through regulating NF-κB/NLRP3 pathway-mediated inflammation and apoptosis. J Recept Signal Transduct 42(2):197–205
Liu J, Rao J, Lou X, Zhai J, Ni Z, Wang X (2018) Upregulated TRIM11 exerts its oncogenic effects in hepatocellular carcinoma through inhibition of P53. Cell Physiol Biochem 44(1):255–266
Zhang ZY, Harischandra DS, Wang R, Ghaisas S, Zhao JY, McMonagle TP et al (2023) TRIM11 protects against tauopathies and is down-regulated in Alzheimer’s disease. Science 381(6656):eadd6696
Ahsan N, Shariq M, Surolia A, Raj R, Khan MF, Kumar P (2024) Multipronged regulation of autophagy and apoptosis: emerging role of TRIM proteins. Cell Mol Biol Lett 29(1):13
Brotto DB, Siena ADD, de Barros II, Carvalho SdCeS, Muys BR, Goedert L et al (2020) Contributions of HOX genes to cancer hallmarks: enrichment pathway analysis and review. Tumor Biol. https://doi.org/10.1177/1010428320918050
Musso G, Gambino R, Tabibian JH, Ekstedt M, Kechagias S, Hamaguchi M et al (2014) Association of non-alcoholic fatty liver disease with chronic kidney disease: a systematic review and meta-analysis. PLoS Med 11(7):e1001680
Zwergal A, Quirling M, Saugel B, Huth KC, Sydlik C, Poli V et al (2006) C/EBPβ blocks p65 phosphorylation and thereby NF-κB-mediated transcription in TNF-tolerant cells. J Immunol 177(1):665–672
Koch DT, Yu H, Beirith I, Schirren M, Drefs M, Liu Y et al (2023) Tigecycline causes loss of cell viability mediated by mitochondrial OXPHOS and RAC1 in hepatocellular carcinoma cells. J Transl Med 21(1):876
Mazgaeen L, Gurung P (2020) Recent advances in lipopolysaccharide recognition systems. Int J Mol Sci 21(2):379
Gallo-Oller G, Ordonez R, Dotor J (2018) A new background subtraction method for Western blot densitometry band quantification through image analysis software. J Immunol Methods 457:1–5
Zhang T, Ma C, Zhang Z, Zhang H, Hu H (2021) NF-κB signaling in inflammation and cancer. MedComm 2(4):618–653
Huang S, Liu D, Sun J, Zhang H, Zhang J, Wang Q et al (2022) Tim-3 regulates sepsis-induced immunosuppression by inhibiting the NF-κB signaling pathway in CD4 T cells. Mol Ther 30(3):1227–1238
Ling X, Wei S, Ling D, Cao S, Chang R, Wang Q et al (2023) Irf7 regulates the expression of Srg3 and ferroptosis axis aggravated sepsis-induced acute lung injury. Cell Mol Biol Lett 28(1):91
Guo Q, Jin Y, Chen X, Ye X, Shen X, Lin M et al (2024) NF-κB in biology and targeted therapy: new insights and translational implications. Signal Transduct Target Ther 9(1):53
Sinha M, Jupe J, Mack H, Coleman TP, Lawrence SM, Fraley SI (2018) Emerging technologies for molecular diagnosis of sepsis. Clin Microbiol Rev. https://doi.org/10.1128/cmr.00089-17
Dai W, Zheng P, Luo D, Xie Q, Liu F, Zhao N et al (2022) LPIN1 is a regulatory factor associated with immune response and inflammation in sepsis. Front Immunol 13:820164
Zhang Q, Bao X, Zhou X, Tang L (2023) Identification and validation of key biomarkers based on RNA methylation genes in sepsis. Front Immunol 14:1231898
Shu Q, She H, Chen X, Zhong L, Zhu J, Fang L (2023) Identification and experimental validation of mitochondria-related genes biomarkers associated with immune infiltration for sepsis. Front Immunol 14:1184126
Wang T, Han J-G, Dong W, Yu Y-H (2024) LCN2 and ELANE overexpression induces sepsis. Medicine 103(7):e37255
Zhao H, Chen Y, Qian L, Du L, Wu X, Tian Y et al (2023) Lycorine protects against septic myocardial injury by activating AMPK-related pathways. Free Radical Biol Med 197:1–14
Zhang N, Ahsan MH, Purchio AF, West DB (2005) Serum amyloid A-luciferase transgenic mice: response to sepsis, acute arthritis, and contact hypersensitivity and the effects of proteasome inhibition. J Immunol 174(12):8125–8134
Ning M, Liu Y, Wang D, Wei J, Hu G, Xing P (2022) Knockdown of TRIM27 alleviated sepsis-induced inflammation, apoptosis, and oxidative stress via suppressing ubiquitination of PPARγ and reducing NOX4 expression. Inflamm Res 71(10):1315–1325
Yang W, Gu Z, Hu H (2020) To TRIM the immunity: from innate to adaptive immunity. Front Immunol 11:552012
Dubey AR, Jagtap YA, Kumar P, Patwa SM, Kinger S, Kumar A et al (2022) Biochemical strategies of E3 ubiquitin ligases target viruses in critical diseases. J Cell Biochem 123(2):161–182
Hannoodee S, Nasuruddin DN (2020) Acute inflammatory response
Matsumoto H, Ogura H, Shimizu K, Ikeda M, Hirose T, Matsuura H et al (2018) The clinical importance of a cytokine network in the acute phase of sepsis. Sci Rep 8(1):13995
Xie Y, Zhao Y, Shi L, Li W, Chen K, Li M et al (2020) Gut epithelial TSC1/mTOR controls RIPK3-dependent necroptosis in intestinal inflammation and cancer. J Clin Investig 130(4):2111–2128
Contarelli S, Fedele V, Melisi D (2020) HOX genes family and cancer: a novel role for homeobox B9 in the resistance to anti-angiogenic therapies. Cancers 12(11):3299
de Bessa Garcia SA, Araújo M, Pereira T, Mouta J, Freitas R (2020) HOX genes function in breast cancer development. Biochim Biophys Acta Rev Cancer 1873(2):188358
Song L, Luo Z-Q (2019) Post-translational regulation of ubiquitin signaling. J Cell Biol 218(6):1776–1786
Zaaroor-Regev D, De Bie P, Scheffner M, Noy T, Shemer R, Heled M et al (2010) Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci 107(15):6788–6793
Yuan R, Wang K, Hu J, Yan C, Li M, Yu X et al (2014) Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying β-catenin degradation. Can Res 74(18):5287–5300
Lingappan K (2018) NF-κB in oxidative stress. Curr Opin Toxicol 7:81–86
Janus P, Szołtysek K, Zając G, Stokowy T, Walaszczyk A, Widłak W et al (2018) Pro-inflammatory cytokine and high doses of ionizing radiation have similar effects on the expression of NF-kappaB-dependent genes. Cell Signal 46:23–31
Lelubre C, Vincent J-L (2018) Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol 14(7):417–427
Riedlinger T, Liefke R, Meier-Soelch J, Jurida L, Nist A, Stiewe T et al (2019) NF-κB p65 dimerization and DNA-binding is important for inflammatory gene expression. FASEB J 33(3):4188
Zingarelli B, Piraino G, Hake PW, O’Connor M, Denenberg A, Fan H et al (2010) Peroxisome proliferator-activated receptor δ regulates inflammation via NF-κB signaling in polymicrobial sepsis. Am J Pathol 177(4):1834–1847
Zhu Y, Xu D (2021) Angiotensin (1–7) attenuates sepsis-induced acute kidney injury by regulating the NF-κB pathway. Front Pharmacol 12:601909
Acknowledgements
None.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
Conception and design of the research: Jiaqi Gan, Fei Pan Acquisition of data: Jiaqi Gan, Fei Pan, Wei Zhang Analysis and interpretation of data: Xiaobing Chen, Zhiyun Qiu Statistical analysis: Jiaqi Gan, Wei Zhang, Zhiyun Qiu Drafting the manuscript: Wei Zhang, Zhiyun Qiu, Jiaqi Gan Revision of manuscript for important intellectual content: Fei Pan, Jiaqi Gan.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2024_10212_MOESM2_ESM.tif
Supplementary file2 (TIF 11999 KB)—Supplementary Figure 1. Modular analysis and diagnostic efficacy of TRIM family genes in the GSE13205 and GSE46955 datasets. (A) Sample dendrogram and trait heat map, different branches represent different GSE13205 dataset samples. (B) Gene dendrogram obtained by average linkage hierarchical clustering. The colored rows below the dendrogram show module assignments determined by dynamic tree cuts. (C) Eigengene adjacency heat map of the correlation between module eigengenes and sample traits. (D and E) Enrichment analysis of 1098 genes in the turquoise module. The x-axis represents the gene ratio; the y-axis represents the GO term or enriched KEGG pathway; the size of the dots represents the odds ratio; the color of the dots represents the level of a p-value. (F) PPI network of TRIM family proteins. Nodes represent proteins or protein domains, while edges represent interactions between these proteins. (G) Four overlapping genes (TRIM11, TRIM33, TRIM27, TRIM42) were expressed in case and control samples of GSE46955 data sets. Red represents the case group, and green represents the control group. (H) ROC curves of TRIM11 in GSE46955 datasets. The horizontal coordinate is a false positive fraction, and the vertical coordinate is a proper positive fraction. GO, Gene Ontology; BP, Biological process; CC, Cellular component; MF, Molecular function; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, Protein-Protein Interaction. ROC, Receiver operating characteristic; AUC: Area under the curve; 95% CI: 95% confidence interval. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
11033_2024_10212_MOESM3_ESM.tif
Supplementary file3 (TIF 1502 KB)—Supplementary Figure 2. Interaction and functional analysis of TRIM11 target genes in the context of sepsis. (A) The UbiBrowser database highlights potential interactions between TRIM11 (center, yellow) and the top 20 target genes. (B) Venn diagram, overlap analysis between TRIM11 target genes and genes downregulated in sepsis (GSE13205 dataset). (C) Analysis of the expression levels of 14 TRIM11 target genes (BTN3A1, EN1, HDAC5, HERC1, HOXA10, HOXA13, HOXA3, HOXB9, ICAM3, LHX6, MAF, SMARCA1, TTN, ZFHX4) in the sepsis group (red) and control group (blue) of the GSE13205 dataset. (D and E) Chord plots of BP and WikiPathway enrichment analysis of TRIM11-related down-regulated genes. The color gradient represents p-values, with darker red reflecting higher significance. BP, Biological process. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gan, J., Zhang, W., Pan, F. et al. TRIM11 modulates sepsis progression by promoting HOXB9 ubiquitination and inducing the NF-κB signaling pathway. Mol Biol Rep 52, 194 (2025). https://doi.org/10.1007/s11033-024-10212-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-10212-5