Abstract
Background
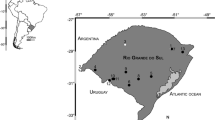
Hybridization associated with polyploidy studies is rare in the tropics. The genus Zygopetalum (Orchidaceae) was investigated here as a case study of Neotropical plants. In the rocky highlands of the Ibitipoca State Park (ISP), southeast Brazil, individuals with intermediate colors and forms between the species Z. maculatum and Z. triste were commonly identified.
Methods and results
Chromosomal analysis and DNA quantity showed a uniform population. Regardless of the aspects related to the color and shape of floral structures, all individuals showed 2n = 96 chromosomes and an average of 14.05 pg of DNA. Irregularities in meiosis associated with chromosome number and C value suggest the occurrence of polyploidy. The genetic distance estimated using ISSR molecular markers revealed the existence of genetic variability not related to morphological clusters. Morphometric measurements of the flower pieces revealed that Z. maculatum shows higher variation than Z. triste although lacking a defined circumscription.
Conclusion
The observed variation can be explained by the polyploid and phenotypic plasticity resulting from the interaction of the genotypes with the heterogeneous environments observed in this habitat.






Similar content being viewed by others
Data availability
Datasets generated during this research are available through the corresponding author upon reasonable request.
References
Wendel JF, Jackson SA, Meyers BC, Wing RA (2016) Evolution of plant genome architecture. Genome Biol 17:37
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100
Albert VA, Barbazuk WB, de Pamphilis CW, Der JP, Leebens-Mack J, Ma H, Palmer JD et al (2013) The Amborella genome and the evolution of flowering plants. Science 342:1241089
Padilla-García N, Šrámková G, Záveská E, Šlenker M, Clo J, Zeisek V, Lučanová M, Rurane I, Kolář F, Marhold K (2023) The importance of considering the evolutionary history of polyploids when assessing climatic niche evolution. J Biogeogr 50:86–100
Rice A, Šmarda P, Novosolov M, Drori M et al (2019) The global biogeography of polyploid plants. Nat Ecol Evol 3:265–273
Flora do Brasil (2020) Jardim Botânico do Rio de Janeiro. http://floradobrasil.jbrj.gov.br/. Acesso em 21 Jan 2022
Oliveira U, Soares-Filho BS, Paglia AP et al (2017) Biodiversity conservation gaps in the Brazilian protected areas. Sci Rep 7:9141
Silveira FAO, Negreiros D, Barbosa NPU et al (2016) Ecology and evolution of plant diversity in the endangered campo rupestre: a neglected conservation priority. Plant Soil 403(1):129–152
Mucina L (2017) Vegetation of Brazilian campos rupestres on siliceous substrates and their global analogues. Flora 238:11–23
Morellato LPC, Silveira FAO (2018) Plant life in campo rupestre: new lessons from an ancient biodiversity hotspot. Flora 238:1–10
Alves RJV, Kolbek J (2010) Can campo rupestre vegetation be floristically delimited based on vascular plant genera? Plant Ecol 207:67–79
Forzza RC, Menini Neto L, Salimena FRG, Zappi D (2013) Capítulo 6: Fanerógamas do Parque Estadual do IBitipoca e suas relações florísticas com outras áreas com campo rupestre de Minas Gerais. In: Forzza RC, Menini Neto L, Salimena FRG, Zappi D (eds) Flora do Parque Estadual do Ibitipoca e seu entorno. Editora UFJF, Juiz de Fora, pp 153–291
Gustafsson ALS, Verola CF, Antonelli A (2010) Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol Biol 10:177
Borba EL, Funch RR, Ribeiro PL, Smidt EC, Silva-Pereira V (2007) Demography, genetic and morphological variability of the endangered Sophronitis sincorana (Orchidaceae) in the Chapada Diamantina, Brazil. Plant Syst Evol 267:129–146
Swartz O (1800) Afhandling om Orchidernes Släegter och deras systematiska indelning. K Vet Akad Handl 21:115–139
Freudenstein JV, Chase MW (2015) Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann Bot 115:665–681
Leal BS, Chaves CJ, Koehler S, Borba EL (2016) When hybrids are not hybrids: a case study of a putative hybrid zone between Cattleya coccinea and C. brevipedunculata (Orchidaceae). Bot J Linn Soc 181:621–639
Stebbins GL (1974) Flowering plants. Evolution above the species level. Arnold, London, p 399
Soltis PS, Marchant DB, Van de Peer Y, Soltis DE (2015) Polyploidy and genome evolution in plants. Curr Opin Genet Dev 35:119–125
Félix LP, Guerra M (2005) Basic chromosome number of terrestrial orchids. Plant Syst Evol 254:131–148
Félix LP, Guerra M (2010) Variation in chromosome number and the basic number of subfamily Epidendroideae (Orchidaceae). Bot J Linn Soc 163:234–278
Hoehne FC (1953) Zygopetalum. Flora Bras 12:1–12
Menini Neto NL, Alves RJV, Barros F, Forzza RC (2007) Orchidaceae do Parque Estadual de Ibitipoca, MG, Brasil. Acta Bot Bras 21:687–696
Rieseberg LH, Ellstrand NC (1993) What can molecular and morphological markers tell us about plant hybridization? CRC Crit Rev Plant Sci 12:213–241
Solano R, Huerta-Espinoza H, Cruz-García G, Ortizriveros F (2019) A new natural hybrid in the genus Laelia (Orchidaceae) from Oaxaca, Mexico. Phytotaxa 402(5):232–240
Scopece G, Palma-Silva C, Cafasso D, Lexer C, Cozzolino S (2020) Phenotypic expression of floral traits in hybrid zones provides insights into their genetic architecture. New Phytol 227:967–975
Yan J, Zhang J, Sun K, Chang D, Bai S, Shen Y, Huang L, Zhang J, Zhang Y, Dong Y (2016) Ploidal level and DNA content of Erianthus arundinaceus as determined by flow cytometry and the association with biological characteristics. PLoS ONE 11:0151948
Turco A, Wagensommer RP, Medagli P, Albano A, D’Emerico S (2024) Advances in the study of Orchidinae subtribe (Orchidaceae) species with 40, 42-chromosomes in the Mediterranean region. Diversity 16(1):41
Trávníček P, Čertner M, Ponert J, Chumová Z, Jersáková J, Suda J (2019) Diversity in genome size and GC content shows adaptive potential in orchids and is closely linked to partial endoreplication, plant life-history traits and climatic conditions. New Phytol 224:1642–1656
Zhang G, Hu Y, Huang MZ, Huang WC, Liu DK, Zhang D, Zhang D, Hu H, Downing JL, Liu Z, Ma H (2023) Comprehensive phylogenetic analyses of Orchidaceae using nuclear genes and evolutionary insights into epiphytism. J Integr Plant Biol 65:1204–1225
GERAIS-CETEC CTDM (1983) Diagnóstico ambiental do Estado de Minas Gerais. Belo Horizonte
Oliveira-Filho AT, Fontes MAL, Viana PL, Valente ASM, Salimena FRG, Ferreira FM (2013) Fanerógamas do Parque Estadual do Ibitipoca e suas relações florísticas com outras áreas com campo rupestre de Minas Gerais. In: Forzza RC, Neto LM, Salimena FRG, Zappi D (eds) Flora do Parque Estadual do Ibitipoca e seu entorno. Editora UFJF, Juiz de Fora, pp 27–52
Rocha GC (2013) Fanerogamas do Parque Estadual do Ibitipoca e suas relações florísticas com outras áreas com campo rupestre de Minas Gerais. In: Forzza RC, Neto LM, Salimena FRG, Zappi D (eds) Flora do Parque Estadual do Ibitipoca e seu entorno. Editora UFJF, Juiz de Fora, pp 27–52
Campacci TVS, Castanho CT, Oliveira RLF, Suzuki RM, Catharino ELM, Koehler S (2017) Effects of pollen origin on apomixis in Zygopetalum mackayi orchids. Flora 226:96–103
Rasband WS (1997) ImageJ, U.S. National Institutes of Health, Bethesda, Maryland, USA. https://imagej.nih.gov/ij/. Accessed June 2020
Hammer O, Harper DAT, Ryan PD (2008) PAST—Palaeontological statistics 1.87 Zuerich, Switzerland
Everitt BS (1978) Graphical techniques for multivariate data. Heinemann Educ. Books Ltd, London
Doležel J, Sgorbati S (1992) Comparison of the three DNA fluorochromes for flow cytometric estimation of nuclear DNA in plants. Physiol Plant 85:625–631
Doležel J, Binarová P, Lucretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31:113–120
Terho P (2013) Flowing software, 2013. www.flowingsoftware.com. Accessed 20 May 2020
Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145
Watanabe K, Yahara T, Denda T, Konsuge K (1999) Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae) statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. J Plant Res 112:45–161
Levan A, Fredga A, Sanderberg AA (1964) Nomenclature for centromeric position in chromosomes. Hereditas 52:201–220
Viccini LF, Pierre PMO, Praça MM et al (2006) Chromosome numbers in the genus Lippia (Verbenaceae). Plant Syst Evol 256:171–178
Williams JGK, Hanafey MK, Rafalski JA, Tingey SV (1993) Genetic analysis using random amplified polymorphic DNA markers. Methods Enzymol 218:704–740
Cruz CD (2016) Genes software—extended and integrated with the R, Matlab and Selegen. Acta Sci Agro 38:547–552
Weir BS (1990) Genetic-data analysis methods for discrete genetic data. Sinauer Assoc. Inc, Sunderland
Anderson JA, Churchill GA, Artique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Bryant D, Moulton V (2004) Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol 21:255–265
Kamvar ZN, Brooks JC, Grünwald NJ (2015) Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet 6:208
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG (2014) Structure plot: a program for drawing elegant structure bar plots in user friendly interface. Springerplus 3:431
Stift M, Kolář F, Meirmans PG (2019) STRUCTURE is more robust than other clustering methods in simulated mixed-ploidy populations. Heredity 123:429–441
Earl DA, Von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goude TJ (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Félix LP, Guerra M (2000) Cytogenetics and cytotaxonomy of some Brazilian species of cymbidioid orchids. Genet Mol Biol 23:957–978
Gomes SSL, Vidal JD, Neves CS, Zorzatto C, Campacci TVS, Lima AK, Koehler S, Viccini LF (2018) Genome size and climate segregation suggest distinct colonization histories of an orchid species from Neotropical high-elevation rocky complexes. Bot J Linn Soc 124:456–465
Leitch IJ, Johnston E, Pellicer J, Hidalgo O, Bennett MD (2019) Plant DNA C-values database. http://www.kew.org/cvalues/. Accessed 26 May 2020
Ehrendorfer F, Krendl F, Habeler E, Sauder W (1968) Chromosome numbers and evolution in primitive angiosperms. Taxon 17:337–468
Stebbins GL (1971) Chromosomal evolution in higher plants. Arnold, London, p 216
Raven P (1975) The bases of angiosperm phylogeny: cytology. Ann Mo Bot Gard 62:724–764
Grant V (1981) Plant speciation. Columbia University Press, New York
Spoelhof JP, Soltis PS, Soltis DE (2017) Pure polyploidy: closing the gaps in autopolyploid research. J Syst Evol 55:340–352
Ramsey J, Schemske DW (2002) Neopolyploidy in flowering plants. Annu Rev Ecol Syst 33:589–639
Buggs RJ, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE (2014) The legacy of diploid progenitors in allopolyploid gene expression patterns. Philos Trans R Soc Lond B 369:20130354
Soltis PS, Soltis DE (2016) Ancient WGD events as drivers of key innovations in angiosperms. Curr Opin Plant Biol 30:159–165
Azevedo CO, Borba EL, van den Berg C (2006) Evidence of natural hybridization and introgression in Bulbophyllum involutum Borba, Semir & F. barros and B. weddellii (Lindl.) Rchb. f. (Orchidaceae) in the Chapada Diamantina, Brazil, by using allozyme markers. Braz J Bot 29:415–421
Moccia MD, Widmer A, Cozzolino S (2007) The strength of reproductive isolation in two hybridizing food-deceptive orchid species. Mol Ecol 16:2855–2866
Conceição AS, Queiroz LP, Borba EL (2008) Natural hybrids in Chamaecrista sect. Absus subsect. Baseophyllum (Leguminosae-Caesalpinioideae): genetic and morphological evidence. Plant Syst Evol 271:19–27
Goulet BE, Roda F, Hopkins R (2017) Hybridization in plants: old ideas, new techniques. Plant Physiol 173:65–78
Tsanakas GF, Mylona PV, Koura K, Gleridou A, Polidoros AN (2018) Genetic diversity analysis of the Greek lentil (Lens culinaris) landrace ‘Eglouvis’ using morphological and molecular markers. Plant Genet Resour 16:469–477
Wu F, Chen J, Wang J, Wang X, Lu Y, Ning Y, Li Y (2019) Intra-population genetic diversity of Buchloedactyloides (Nutt.) Engelm (buffalo grass) determined using morphological traits and sequence-related amplified polymorphism markers. 3 Biotech 9:97
Pinheiro LR, Rabbani ARC, da Silva AVC, da Silva LA, Pereira KLG, Diniz LEC (2012) Genetic diversity and population structure in the Brazilian Cattleya labiata (Orchidaceae) using RAPD and ISSR markers. Plant Syst Evol 298:1815–1825
Pinheiro F, Cardoso-Gustavson P, Suzuki RM, Abrão MCR, Guimarães LRS, Draper D, Moraes AP (2015) Strong post-zygotic isolation prevents introgression between two hybridizing neotropical orchids Epidendrum denticulatum and E. fulgens. Evol Ecol 29:229–248
Rodrigues LA, Paiva Neto VBD, Boaretto AG, Oliveira JFD, Torrezan MDA, Lima SFD, Otoni WC (2015) In vitro propagation of Cyrtopodium saintlegerianum rchb. f. (orchidaceae), a native orchid of the Brazilian savannah. Crop Breed Appl Biotechnol 15:10–17
Li A, Ge S (2006) Genetic variation and conservation of Changnienia amoena, an endangered orchid endemic to China. Plant Syst Evol 258:251–260
Azizi MMF, Lau HY, Abu-Bakar N (2021) Integration of advanced technologies for plant variety and cultivar identification. J Biosci 46:91. https://doi.org/10.1007/s12038-021-00214-x
Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP (2010) Computational solutions to large-scale data management and analysis. Nat Rev Genet 11:647–657
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Mucina L (2018) Vegetation of Brazilian campos rupestres on siliceous substrates and their global analogues. Flora 238:11–23
Lambrecht SC, Dawson TE (2007) Correlated variation of floral and leaf traits along a moisture availability gradient. Oecologia 151:574–583
Pavarese G, Tranchida-Lombardo V, Galesi R, D’emerico S, Casotti R, Cristaudo A, Cozzolino S (2013) When polyploidy and hybridization produce a fuzzy taxon: the complex origin of the insular neoendemic Neotinea commutate (Orchidaceae). Bot J Linn Soc 173:707–720
Gratani L (2014) Plant phenotypic plasticity in response to environmental factors. Adv Bot 2014:17
Soltis DE, Soltis PS, Schemske DW, Hancock JF, Thompson JN, Husband BC, Judd WS (2007) Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56:13–30
Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA (2016) On the relative abundance of autopolyploids and allopolyploids. New Phytol 210:391–398
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501
Ackerman JD, Morales M, Tremblay R (2011) Darwin’s orchids: their variation, plasticity, and natural selection. Lankesteriana 11:179–184
Moreira ASFP, Borba EL, Lemos-Filho JP (2013) Testing arbitrary classes of light in a physiognomically heterogeneous area of ‘campo rupestre’ vegetation. An Acad Bras Cienc 85:635–648
Viccini LF, Silveira RS, do Vale AA et al (2014) Citral and linalool content has been correlated to DNA content in Lippia alba (Mill.) N.E. Brown (Verbenaceae). Ind Crops Prod 59:14–19
Acknowledgements
The authors wish to thank Instituto Estadual de Florestas (IEF) for making the collection of the samples possible.
Funding
This work was financially supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais-Fapemig (APQ-02096-14, APQ-02348-21,), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Capes and Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (314.443/2021-5).
Author information
Authors and Affiliations
Contributions
SSLG, LMN, and LFV designed the research; SSLG, JMLL, EMM, and EGC performed the research with the support of ALSA, MAM, JMSC, LMN, and LFV. SSLG, JMLL, EMM, ALSA, MAM, JMSC, LMN, and LFV analyzed the data; SSLG, JMLL, EMM, and LFV wrote the article with input from all the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gomes, S.S.L., Lopes, J.M.L., de Matos, E.M. et al. Phenotypic variation seems not to be associated with the genetic profile in Zygopetalum (Orchidaceae): a case study of a high-elevation rocky complex. Mol Biol Rep 51, 582 (2024). https://doi.org/10.1007/s11033-024-09528-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09528-z