Abstract
Background
A novel lytic bacteriophage (phage) was isolated with Pseudomonas mendocina strain STP12 (P. mendocina) from the untreated site of Sewage Treatment Plant of Lovely Professional University, India. P. mendocina is a Gram-negative, rod-shaped, aerobic bacterium belonging to the family Pseudomonadaceae and has been reported in fifteen (15) cases of economically important diseases worldwide.
Methods and results
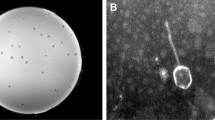
Here, a novel phage specifically infecting and killing P. mendocina strain STP12 was isolated from sewage sample using enrichment, spot test and double agar overlay (DAOL) method and was designated as vB_PmeS_STP12. The phage vB-PmeS-STP12 was viable at wide range of pH and temperature ranging from 4 to10 and − 20 to 70 °C respectively. Host range and efficiency of plating (EOP) analysis indicated that phage vB-PmeS-STP12 was capable of infecting and killing P. mendocina strain STP6 with EOP of 0.34. Phage vB_PmeS_STP12 was found to have a significant bacterial reduction (p < 0.005) at all the doses administered, particularly at optimal MOI of 1 PFU/CFU, compared to the control. Morphological analysis using high resolution transmission electron microscopy (HR-TEM) revealed an icosahedral capsid of ~ 55 nm in diameter on average with a short, non-contractile tail. The genome of vB_PmeS_STP12 is a linear, dsDNA containing 36,212 bp in size with a GC content of 58.87% harbouring 46 open reading frames (ORFs). The 46 predicted ORFs encode proteins with functional information categorized as lysis, replication, packaging, regulation, assembly, infection, immune, and hypothetical. However, the genome of vB_PmeS_STP12 appeared to be devoid of tRNAs, integrase gene, toxins genes, virulence factors, antimicrobial resistance genes (ARGs) and CRISPR arrays. The blast analysis with phylogeny revealed that vB_PmeS_STP12 is genetically similar to Pseudomonas phage PMBT14, Pseudomonas phage Almagne and Serratia phage Serbin with a highest identity of 74.00%, 74.93% and 59.48% respectively.
Conclusions
Taken together, characterization, morphological analysis and genome-informatics indicated that vB_PmeS_STP12 is podovirus morphotype belonging to the class Caudoviticetes, family Zobellviridae which appeared to be devoid of integrase gene, ARGs, CRISPR arrays, virulence factors and toxins genes, exhibiting stability and infectivity at wide range of pH (4 to10) and temperature (–20 to 70 °C), thereby making vB_PmeS_STP12 suitable for phage therapy or biocontrol. Based on the bibliometric analysis and data availability with respect to sequences deposited in GenBank, this is the first report of a phage infecting Pseudomonas mendocina.






Similar content being viewed by others
Data availability
The genome sequence of Pseudomonas mendocina phage vB_PmeS_STP12 and the 16 S rRNA sequence of its host Pseudomonas mendocina strain STP12 have been deposited in the GenBank under the accession numbers OR754314 and OR188352 respectively (https://www.ncbi.nlm.nih.gov/nuccore). The 16 S rRNA sequences of the alternative hosts used for host range analysis namely P. mendocina strain STP6, S. stutzeri strain STP9, B. stratosphericus strain STP9 and P. mirabilis strain STP14 have also been deposited in the GenBank under the accession numbers OR188127, OR188486, OR186294, and OR187366 respectively (https://www.ncbi.nlm.nih.gov/nuccore).
Change history
05 May 2024
A Correction to this paper has been published: https://doi.org/10.1007/s11033-024-09536-z
Abbreviations
- AMR:
-
Antimicrobial resistance
- Anti-CRISPRdb:
-
Anti- Clustered regularly interspaced short palindromic repeats database
- Anti-CRISPR proteins:
-
Anti- Clustered regularly interspaced short palindromic repeats proteins
- ARGs:
-
Antimicrobial resistance genes
- BLASTn:
-
Basic local alignment search tool for nucleotide
- BLASTP:
-
Basic local alignment search tool for proteins
- BOD:
-
Biochemical oxygen demand
- CARD:
-
Comprehensive antibiotic research database
- CIF:
-
Central instrumentation facility
- COD:
-
Chemical oxygen demand
- CRISPR-arrays:
-
Clustered regularly interspaced short palindromic repeats-arrays
- CRISPRCasFinder:
-
Clustered regularly interspaced short palindromic repeats-CRISPR-associated protein finder
- DAOL:
-
Double agar overlay
- DHF:
-
Dihydrofolate
- DNA:
-
Deoxyribonucleic acid
- DO:
-
Dissolved oxygen
- DSBs:
-
DNA double-strand breaks
- dsDNA:
-
Double-stranded DNA
- dUMP:
-
2’-deoxyuridine- 5’-monophosphate
- dTMP:
-
2’-deoxythymidine-5’-monophosphate
- eggNOG:
-
Evolutionary genealogy of genes:Non- supervised Orthologous Group
- EC:
-
Electrical conductivity
- EPA:
-
Environmental protection agency
- HR-TEM:
-
High resolution-transmission electron microscopy
- HQ:
-
High quality
- HiFi:
-
High fidelity
- ICTV:
-
International Committee on Taxonomy of Viruses
- IISER:
-
Indian institute of science research and education
- LB:
-
Luria Bertani
- LPU:
-
Lovely Professional University
- MDR Bacteria:
-
Multidrug-resistant bacteria
- MMseqs:
-
Many-against-Many sequence searching
- MOI:
-
Multiplicity of infection
- mTHF:
-
5,10-methylenetetrahydrofolate
- NCBI:
-
National Center for Biotechnology Information
- NT:
-
Nucleotide
- NR:
-
Non-redundant
- ON:
-
Overnight
- ORFs:
-
Open reading frames
- PCR:
-
Polymerase chain reaction
- PFU:
-
Plaque forming unit
- PE:
-
Pair-end
- RNA:
-
Ribonucleic acid
- rRNA:
-
ribosomal RNA
- SM buffer:
-
Sodium magnesium buffer
- ssDNA:
-
Single-stranded DNA
- STP:
-
Sewage treatment plant
- TDS:
-
Total dissolved solid
- TEM:
-
Transmission electron microscopy
- TMHMM:
-
Transmembrane hidden Markov model
- tRNA:
-
Transfer RNA
- tRNAscan-SE:
-
Transfer RNA scan in genomic sequence
- TSS:
-
Total suspended solid
- US:
-
United States
- UV light:
-
Ultraviolet light
- VFDB:
-
Virulence factor database
- WGS:
-
Whole-genome sequencing
References
Usman SS, Uba AI, Christina E (2023) Bacteriophage genome engineering for phage therapy to combat bacterial antimicrobial resistance as an alternative to antibiotics. Mol Biol Rep 50(8):7055–7067. https://doi.org/10.1007/s11033-023-08557-4
Keller M, Wellington A, De Souza V, Capua J, Novello DL, Sillankorva S (2023) Complete genome sequence of a novel bacteriophage vB _ Pci _ PCMW57 infecting phytobacteria Pseudomonas cichorii, pp. 7105–7111
Lamichhane B et al (2024) Salmonellosis: an overview of Epidemiology, Pathogenesis, and innovative approaches to mitigate the Antimicrobial resistant infections, pp. 1–55
Kordi M, Talkhounche PG, Vahedi H, Farrokhi N, Tabarzad M (2024) Heterologous production of antimicrobial peptides: notes to consider. Protein J no November. https://doi.org/10.1007/s10930-023-10174-w
Lucchino EC, Gennaro G, Favoino F, Goia F (2022) Jo ur na l P r f. Build Environ 109704. https://doi.org/10.1016/j.xplc.2024.100812
Sen D, Mukhopadhyay P (2024) Antimicrobial Resistance (AMR) management using CRISPR-Cas based genome editing. Gene Genome Ed 100031. https://doi.org/10.1016/j.ggedit.2024.100031
Sreekumaran S et al (2024) Predicting Human Risk with Multidrug Resistant Enterobacter hormaechei MS2 having MCR 9 Gene isolated from the feces of healthy broiler through whole-genome sequence-based analysis. Curr Microbiol 81(1):1–8. https://doi.org/10.1007/s00284-023-03492-w
Wasan H, Reeta KH, Gupta YK (2023) Strategies to improve antibiotic access and a way forward for lower middle-income countries. J Antimicrob Chemother 79(1):1–10. https://doi.org/10.1093/jac/dkad291
Collaborators AR (2022) Articles Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. 6736(21). https://doi.org/10.1016/S0140-6736(21)02724-0
Online VA et al (2024) Materials advances delivery and enhanced therapeutic effects. https://doi.org/10.1039/d3ma00817g
Mustafa MI, Mohammed A (2023) Developing recombinant antibodies by phage display technology to neutralize viral infectious diseases, SLAS Discov, no. December pp. 1–8, 2024, https://doi.org/10.1016/j.slasd.2024.01.001
Helmy YA et al (2023) Antimicrobial Resistance and recent Alternatives to Antibiotics for the Control of Bacterial Pathogens with an emphasis on Foodborne pathogens
Zahedipour F, Zahedipour F, Zamani P, Reza M, Sahebkar A (2023) Harnessing CRISPR technology for viral therapeutics and vaccines: from preclinical studies to clinical applications, Virus Res, vol. 341, no. December p. 199314, 2024, https://doi.org/10.1016/j.virusres.2024.199314
Grigson SR, Giles SK, Edwards RA, Papudeshi B (2023) Knowing and naming: phage annotation and nomenclature for phage therapy. Clin Infect Dis 77:352–359. https://doi.org/10.1093/cid/ciad539
Aragone MDR, Maurizi DM, Clara LO, Estrada JLN, Ascione A (1992) Pseudomonas mendocina, an environmental bacterium isolated from a patient with human infective endocarditis. J Clin Microbiol 30(6):1583–1584. https://doi.org/10.1128/jcm.30.6.1583-1584.1992
Gani M, Rao S, Miller M, Scoular S (2019) Pseudomonas Mendocina bacteremia: a case study and review of literature. Am J Case Rep 20:453–458. https://doi.org/10.12659/AJCR.914360
Rapsinski GJ, Makadia J, Bhanot N, Min Z (2016) Pseudomonas mendocina native valve infective endocarditis: a case report. J Med Case Rep 6–10. https://doi.org/10.1186/s13256-016-1057-6
Gani M, Miller M, Scoular S (2019) Pseudomonas Mendocina Bacteremia: a Case Study and Review of Literature. 453–458. https://doi.org/10.12659/AJCR.914360
Ioannou P (2020) A Systematic Review of Human Infections by Pseudomonas mendocina, no. January, pp. 1–9
Sutter ST, Halter J, Frei R, Widmer AF (2011) Pseudo-outbreak of Pseudomonas mendocina in stem cell cultures. J Hosp Infect 77(1):70–72. https://doi.org/10.1016/j.jhin.2010.08.008
Greenough N, Gerry D (2021) Clinical infection in Practice First UK case report of a patient with Pseudomonas mendocina bacteraemia. Clin Infect Pract 10:100065. https://doi.org/10.1016/j.clinpr.2021.100065
Bosco K, Lynch S, Sandaradura I, Khatami A (2023) Therapeutic phage monitoring: a review. Clin Infect Dis 77:384–394. https://doi.org/10.1093/cid/ciad497
Yerushalmy O et al (2023) Towards standardization of Phage susceptibility testing: the Israeli phage therapy Center ‘ clinical phage Microbiology ’— A Pipeline proposal. Clin Infect Dis 77:337–351. https://doi.org/10.1093/cid/ciad514
Fajardo-lubian A, Venturini C (2023) Use of bacteriophages to target intracellular pathogens. Clin Infect Dis 77:423–432. https://doi.org/10.1093/cid/ciad515
Suh GA, Ferry T, Abdel MP (2023) Phage therapy as a Novel Therapeutic for the treatment of bone and joint infections. Clin Infect Dis 77:407–415. https://doi.org/10.1093/cid/ciad533
Malik DJ et al (2023) Advanced Manufacturing, Formulation and Microencapsulation of Therapeutic Phages. Clin Infect Dis 77:370–383. https://doi.org/10.1093/cid/ciad555
Abdelsattar AS, Dawoud A, Makky S, Nofal R, Aziz RK, El-Shibiny A (2021) Bacteriophages: from isolation to application. Curr Pharm Biotechnol 22. https://doi.org/10.2174/1389201022666210426092002
Brives C, Pourraz J (2020) Phage therapy as a potential solution in the fight against AMR: obstacles and possible futures. Palgrave Commun 6(1):1–11. https://doi.org/10.1057/s41599-020-0478-4
Ji M, Liu Z, Sun K, Li Z, Fan X, Li Q (2021) Bacteriophages in water pollution control: advantages and limitations. Front Environ Sci Eng 15(5). https://doi.org/10.1007/s11783-020-1378-y
Abedon ST (2023) How simple Maths can inform our Basic understanding of phage therapy. Clin Infect Dis 77:401–406. https://doi.org/10.1093/cid/ciad480
Hyman P (2019) Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals 12(1). https://doi.org/10.3390/ph12010035
Abdelsattar AS, Safwat A, Nofal R, Elsayed A, Makky S, El-Shibiny A (2021) Isolation and characterization of bacteriophage ZCSE6 against Salmonella spp.: phage application in milk. Biologics 1(2):164–176. https://doi.org/10.3390/biologics1020010
Ali S, Karaynir A, Salih H, Oncü S, Bozdo B (2022) Characterization, genome analysis and antibiofilm efficacy of lytic Proteus phages RP6 and RP7 isolated from university hospital sewage, vol. 326, no. October 2023, https://doi.org/10.1016/j.virusres.2023.199049
Sevcik K, Preston P, Aulner M, Noordewier B, Tolsma SS (2023) Complete Genome Sequences of Cluster S Mycobacteriophages Beelzebub, Raela, and RedRaider77. Microbiol Resour Announc 12(1):17–19. https://doi.org/10.1128/mra.01173-22
Mardiana M, Teh SH, Tsai YC, Yang HH, Lin LC, Lin NT (2023) Characterization of a novel and active temperate phage vB_AbaM_ABMM1 with antibacterial activity against Acinetobacter baumannii infection. Sci Rep 13(1):1–13. https://doi.org/10.1038/s41598-023-38453-7
Loh B et al (2021) Complete Genome Sequences of Bacteriophages Kaya, Guyu, Kopi, and TehO, which target clinical strains of Pseudomonas aeruginosa. Microbiol Resour Announc 10(48):14–16. https://doi.org/10.1128/mra.01043-21
Kazimierczak J, Wójcik EA, Witaszewska J, Guzi A (2019) Complete genome sequences of Aeromonas and Pseudomonas phages as a supportive tool for development of antibacterial treatment in aquaculture, pp. 1–12
Guo J, Xu S, Liu Y, Zhang C, Hou S (2023) Complete genome sequence of Stutzerimonas stutzeri strain SOCE 002, a Marine Bacterium isolated from the Surface seawater of Dapeng Bay. Microbiol Resour Announc 12(5):10–12. https://doi.org/10.1128/mra.00150-23
Akmal M, Nishiki I, Zrelovs N, Yoshida T (2022) Complete genome sequence of a novel lytic bacteriophage, PLG – II, specific for Lactococcus garvieae serotype II strains that are pathogenic to fish. Arch Virol 167(11):2331–2335. https://doi.org/10.1007/s00705-022-05568-7
Kwon H et al (2024) Genomic and biological characteristics of a novel lytic phage, vB_MscM-PMS3, infecting the opportunistic zoonotic pathogen Mammaliicoccus Sciuri. Arch Virol 169(1):1–6. https://doi.org/10.1007/s00705-023-05940-1
Mareso C et al (2023) Optimization of long-range PCR protocol to prepare filaggrin exon 3 libraries for PacBio long-read sequencing. Mol Biol Rep 50(4):3119–3127. https://doi.org/10.1007/s11033-022-08170-x
Maffei E et al (2024) Phage paride can kill dormant, antibiotic-tolerant cells of Pseudomonas aeruginosa by direct lytic replication. Nat Commun 15(1):175. https://doi.org/10.1038/s41467-023-44157-3
Fredriksson NJ, Hermansson M, Wilén BM (2013) The choice of PCR primers has great impact on assessments of Bacterial Community Diversity and Dynamics in a Wastewater Treatment Plant. PLoS ONE 8(10):1–20. https://doi.org/10.1371/journal.pone.0076431
Galkiewicz JP, Kellogg CA (2008) Cross-kingdom amplification using Bacteria-specific primers: complications for studies of coral microbial ecology. Appl Environ Microbiol 74(24):7828–7831. https://doi.org/10.1128/AEM.01303-08
Miyashita A et al (2009) Development of 16S rRNA gene-targeted primers for detection of archaeal anaerobic methanotrophs (ANMEs). FEMS Microbiol Lett 297(1):31–37. https://doi.org/10.1111/j.1574-6968.2009.01648.x
Salim A et al (2021) Bacteriophage-based control of biogenic hydrogen sulphide produced by multidrug resistant Salmonella enterica in synthetic sewage. J Environ Chem Eng 9(4):105797. https://doi.org/10.1016/j.jece.2021.105797
Dewanggana MN, Evangeline C, Ketty MD, Waturangi DE, Yogiara, Magdalena S (2022) Isolation, characterization, molecular analysis and application of bacteriophage DW-EC to control enterotoxigenic Escherichia coli on various foods. Sci Rep 12(1):1–10. https://doi.org/10.1038/s41598-021-04534-8
Salim A, Madhavan A, Subhash S, Prasad M, Nair BG, Pal S (2022) Escherichia coli ST155 as a production-host of three different polyvalent phages and their characterisation with a prospect for wastewater disinfection. Sci Rep 12(1):1–16. https://doi.org/10.1038/s41598-022-24134-4
Ma Y et al (2016) Isolation and molecular characterisation of Achromobacter phage phiAxp-3, an N4-like bacteriophage, Sci. Rep, vol. 6, no. February, pp. 1–12, https://doi.org/10.1038/srep24776
Nivas D, Ramesh N, Krishnakumar V, Rajesh P, King Solomon E, Kannan VR (2015) Distribution, isolation and characterization of lytic bacteriophages against multi-drug resistant and extended-spectrum of β-lactamase producing pathogens from hospital effluents. Asian J Pharm Clin Res 8(2):384–389
Runa V, Wenk J, Bengtsson S, Jones BV, Lanham AB (October, 2021) Bacteriophages in Biological Wastewater Treatment systems: occurrence, characterization, and function. Front Microbiol 12. https://doi.org/10.3389/fmicb.2021.730071
Manohar P, Tamhankar AJ, Lundborg CS, Ramesh N (2018) Isolation, characterization and in vivo efficacy of Escherichia phage myPSH1131. PLoS ONE 13(10):1–17. https://doi.org/10.1371/journal.pone.0206278
Porayath C et al (2018) Characterization of the bacteriophages binding to human matrix molecules. Int J Biol Macromol 110:608–615. https://doi.org/10.1016/j.ijbiomac.2017.12.052
Czajkowski R, Ozymko Z, Lojkowska E (2016) Application of zinc chloride precipitation method for rapid isolation and concentration of infectious Pectobacterium spp. and Dickeya spp. lytic bacteriophages from surface water and plant and soil extracts. 29–33. https://doi.org/10.1007/s12223-015-0411-1
Santos MA (1991) An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. 19(19):5442
Glonti T, Pirnay J (1940) In Vitro Measurement Of Phage Characteristics That Are Im- portant for Phage Therapy Outcome, Viruses, vol. 14, no. pp. 1–23, 2022
Mankovich AG, Maciel K, Kavanaugh M, Kistler E, Muckle E, Weingart CL (2023) Phage-antibiotic synergy reduces Burkholderia cenocepacia population. BMC Microbiol 23(1):1–12. https://doi.org/10.1186/s12866-022-02738-0
Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R (2019) Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, klebsiella pneumoniae, and enterobacter species, Front. Microbiol, vol. 10, no. MAR, pp. 1–12, https://doi.org/10.3389/fmicb.2019.00574
Cieplak T, Soffer N, Sulakvelidze A, Nielsen DS (2018) A bacteriophage cocktail targeting Escherichia coli reduces E. Coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes 9(5):391–399. https://doi.org/10.1080/19490976.2018.1447291
Sun WJ et al (2012) A novel bacteriophage KSL-1 of 2-Keto-gluconic acid producer Pseudomonas fluorescens K1005: isolation, characterization and its remedial action. BMC Microbiol 12:1–8. https://doi.org/10.1186/1471-2180-12-127
Nayfach S, Camargo AP, Schulz F, Eloe-fadrosh E, Roux S, Kyrpides NC (May, 2021) Metagenome-assembled viral genomes. Nat Biotechnol 39. https://doi.org/10.1038/s41587-020-00774-7
Hyatt D, Chen G, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification
Table S, Mmseqs F, Fig S (2007) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. 1–3. https://doi.org/10.1038/nbt.3988
Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL (2000) Prediction of transcription terminators in bacterial genomes. 27–33. https://doi.org/10.1006/jmbi.2000.3836
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. 25(5):955–964
Krogh A, Larsson È, Von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov Model: application to complete genomes. https://doi.org/10.1006/jmbi.2000.4315
Couvin D et al (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins, vol. 46, no. May, pp. 246–251, https://doi.org/10.1093/nar/gky425
Dong C et al (2017) September., Anti-CRISPRdb: a comprehensive online resource for anti-CRISPR proteins, vol. 46, no. pp. 393–398, 2018, https://doi.org/10.1093/nar/gkx835
Li Y, Bondy-denomy J (2020) Review Anti-CRISPRs go viral: the infection biology of CRISPR-Cas inhibitors. Cell Host Microbe 29(5):704–714. https://doi.org/10.1016/j.chom.2020.12.007
Eitzinger S et al (2020) Machine learning predicts new anti-CRISPR proteins. 48(9):4698–4708. https://doi.org/10.1093/nar/gkaa219
Chen L et al (2005) VFDB: a reference database for bacterial virulence factors. 33:325–328. https://doi.org/10.1093/nar/gki008
Mcarthur AG et al (2013) The Comprehensive Antibiotic Resistance Database, vol. 57, no. 7, pp. 3348–3357, https://doi.org/10.1128/AAC.00419-13
Buchfink B, Reuter K, Drost HG (2021) Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18(4):366–368. https://doi.org/10.1038/s41592-021-01101-x
Saitou N, Nei M (1987) The neighbor-joining method: a New Method for reconstructing phylogenetic trees ’. 4(4):406–425
Wang RH et al (2023) Phag eScope: a w ell-annotat ed bact eriophag e database with automatic analyses and visualizations, pp. 1–6
Tisza MJ, Buck CB (2021) A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. 33:1–11. https://doi.org/10.1073/pnas.2023202118
Turner D, Kropinski AM, Adriaenssens EM (2021) A roadmap for genome-based phage taxonomy. Viruses 13(3):1–10. https://doi.org/10.3390/v13030506
Moraru C, Nayfach S, Lab B (2022) and U. States Vicente Pérez-Brocal, Phage family classification under Caudoviricetes: A review of current tools using the latest ICTV classification framework, Front. Microbiol, vol. 13, p. 1032186, [Online]. Available: https://doi.org/10.3389/fmicb.2022.1032186
Turner D, Shkoporov AN, Lood C, Millard AD, Dutilh BE (2023) Abolishment of morphology – based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch Virol 168(2):1–9. https://doi.org/10.1007/s00705-022-05694-2
Evseev P, Gutnik D, Shneider M, Miroshnikov K (2023) Use of an Integrated Approach Involving AlphaFold predictions for the Evolutionary Taxonomy of Duplodnaviria viruses. Biomolecules 13(1). https://doi.org/10.3390/biom13010110
Grami E, Laadouze I, Ben Tiba S, Hafiane A, Sealey KS, Saidi N (2023) Isolation, characterization, and comparative genomic analysis of vB_Pd_C23, a Novel bacteriophage of Pantoea dispersa. Curr Microbiol 80(1):1–11. https://doi.org/10.1007/s00284-022-03152-5
Gorodnichev RB et al (2023) Isolation and characterization of the First Zobellviridae Family Bacteriophage infecting Klebsiella pneumoniae. Int J Mol Sci 24(4). https://doi.org/10.3390/ijms24044038
Krupovic M et al (2021) Bacterial Viruses Subcommittee and Archaeal Viruses Subcommittee of the ICTV: update of taxonomy changes in 2021. Arch Virol 166(11):3239–3244. https://doi.org/10.1007/s00705-021-05205-9
Zheng K et al (2023) Genomic diversity and ecological distribution of marine Pseudoalteromonas phages. Mar Life Sci Technol 5(2):271–285. https://doi.org/10.1007/s42995-022-00160-z
Yang M et al (2023) Genomic diversity and biogeographic distributions of a novel lineage of bacteriophages that infect marine OM43 bacteria. Microbiol Spectr 11(5). https://doi.org/10.1128/spectrum.04942-22
Ge F et al (2023) Characterization and genomic analysis of Stutzerimonas stutzeri phage vB _ PstS _ ZQG1, representing a novel viral genus. Virus Res 336:199226. https://doi.org/10.1016/j.virusres.2023.199226. June
Koberg S, Gieschler S, Brinks E, Wenning M, Neve H, Franz CMAP (2018) Genome sequence of the novel virulent bacteriophage PMBT14 with lytic activity against Pseudomonas fluorescens DSM 50090R. Arch Virol 163(9):2575–2577. https://doi.org/10.1007/s00705-018-3882-y
Cantalapiedra CP, Huerta-cepas J, Hern A, Letunic I, Bork P (2021) eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale, vol. 38, no. 12, pp. 5825–5829, https://doi.org/10.1093/molbev/msab293
McMillan S, Edenberg HJ, Radany EH, Friedberg RC, Friedberg EC (1981) Den V gene of bacteriophage T4 codes for both pyrimidine dimer-DNA glycosylase and apyrimidinic endonuclease activities. J Virol 40(1):211–223. https://doi.org/10.1128/jvi.40.1.211-223.1981
Radany EH, Nguyen HT, Minton KW (1987) Activities involved in base excision repair of bacteriophage T4 and lambda DNA in vivo. MGG Mol Gen Genet 209(1):83–89. https://doi.org/10.1007/BF00329840
Chang Y, Wang Q, Su T, Qi Q (2021) Identification of phage recombinase function unit in Genus Corynebacterium. Appl Microbiol Biotechnol 105(12):5067–5075. https://doi.org/10.1007/s00253-021-11384-x
Huerta-Cepas J et al (2019) EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47 D1, pp. D309–D314. https://doi.org/10.1093/nar/gky1085
Acknowledgements
We would like to thank Lovely Professional University for providing the facilities to carry out the present study; Associate Professor Gabriel Magno de Freitas Almeida of UiT The Arctic University of Norway for his constructive guidance; the CIF, IISER Thiruvananthapuram for conducting the HR-TEM analysis.
Funding
No funding is received.
Author information
Authors and Affiliations
Contributions
Sani Sharif Usman: Conceptualization, Formal analysis, Investigation, Project administration, Software, Writing – original draft, Writing – review & editing. Evangeline Christina: Conceptualization, Formal analysis, Investigation, Project administration, Software, Supervision, Writing – original draft, Writing – review & editing. The authors read and approved the final manuscript as well as agreed to authorship and submission of the manuscript for publication.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors have declared that no competing interest exists.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The article title is corrected as ‘Characterization and genome-informatic analysis of a novel lytic Pseudomonas mendocina phage vB_PmeS_STP12 suitable for phage therapy or biocontrol’
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Usman, S.S., Christina, E. Characterization and genome-informatic analysis of a novel lytic Pseudomonas mendocina phage vB_PmeS_STP12 suitable for phage therapy or biocontrol. Mol Biol Rep 51, 419 (2024). https://doi.org/10.1007/s11033-024-09362-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09362-3