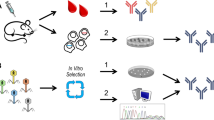
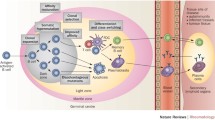
Abstract
Synthetic antibodies (Abs) are a class of engineered proteins designed to mimic the functions of natural Abs. These are produced entirely in vitro, eliminating the need for an immune response. As such, synthetic Abs have transformed the traditional methods of raising Abs. Likewise, deep sequencing technologies have revolutionized genomics and molecular biology. These enable the rapid and cost-effective sequencing of DNA and RNA molecules. They have allowed for accurate and inexpensive analysis of entire genomes and transcriptomes. Notably, via deep sequencing it is now possible to sequence a person’s entire B-cell receptor immune repertoire, termed BCR sequencing. This procedure allows for big data explorations of natural Abs associated with an immune response. Importantly, the identified sequences have the ability to improve the design and engineering of synthetic Abs by offering an initial sequence framework for downstream optimizations. Additionally, machine learning algorithms can be introduced to leverage the vast amount of BCR sequencing datasets to rapidly identify patterns hidden in big data to effectively make in silico predictions of antigen selective synthetic Abs. Thus, the convergence of BCR sequencing, machine learning, and synthetic Ab development has effectively promoted a new era in Ab therapeutics. The combination of these technologies is driving rapid advances in precision medicine, diagnostics, and personalized treatments.

Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- Abs:
-
Antibodies
- mAbs:
-
Monoclonal Abs
- NGS:
-
Next-generation sequencing
- BCRs:
-
B-cell receptors
- Ig:
-
Immunoglobulins
- CDRs:
-
Complementary determining regions
- V:
-
Variable
- J:
-
Joining
References
Bradbury ARM, Sidhu SS, Dübel S et al (2011) Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 29:245–254. https://doi.org/10.1038/nbt.1791
Adams JJ, Sidhu SS (2014) Synthetic antibody technologies. Curr Opin Struct Biol 24:1–9. https://doi.org/10.1016/j.sbi.2013.11.003
Chen G, Sidhu SS (2014) Design and generation of synthetic antibody libraries for phage display. Methods Mol Biol 1131:113–131. https://doi.org/10.1007/978-1-62703-992-5_8
Chen G, Gorelik L, Simon KJ et al (2015) Synthetic antibodies and peptides recognizing progressive multifocal leukoencephalopathyspecific point mutations in polyomavirus JC capsid viral protein 1. MAbs 7:681–692. https://doi.org/10.1080/19420862.2015.1038447
Miersch S, Sidhu SS (2012) Synthetic antibodies: concepts, potential and practical considerations. Methods 57:486–498. https://doi.org/10.1016/j.ymeth.2012.06.012
Marks JD, Hoogenboom HR, Bonnert TP et al (1991) By-passing immunization. J Mol Biol 222:581–597. https://doi.org/10.1016/0022-2836(91)90498-U
Sidhu SS, Fellouse FA (2006) Synthetic therapeutic antibodies. Nat Chem Biol 2:682–688. https://doi.org/10.1038/nchembio843
Fuh G (2007) Synthetic antibodies as therapeutics. Expert Opin Biol Ther 7:73–87. https://doi.org/10.1517/14712598.7.1.73
Kimball JW (2011) Monoclonal Antibodies. Online text B 1131:447–476. https://doi.org/10.1385/1-59259-049-7:233
Zola H (2013) Monoclonal antibodies. CRC Press, Boca Raton
Kipriyanov SM (2009) Antibody phage display. Antib Phage Disp 562:177–193. https://doi.org/10.1007/978-1-60327-302-2
Hanes J, Jermutus L, Weber-Bornhauser S et al (1998) Ribosome display efficiently selects and evolves high-affinity antibodies in vitro from immune libraries. Proc Natl Acad Sci 95:14130–14135. https://doi.org/10.1073/pnas.95.24.14130
Sergeeva A, Kolonin MG, Molldrem JJ et al (2006) Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev 58:1622–1654. https://doi.org/10.1016/j.addr.2006.09.018
Boder ET, Wittrup KD (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol 15:553–557. https://doi.org/10.1038/nbt0697-553
Deantonio C, Cotella D, Macor P, et al (2014) Human Monoclonal Antibodies. 1060:277–295. https://doi.org/10.1007/978-1-62703-586-6
Fellouse FA, Esaki K, Birtalan S et al (2007) High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol 373:924–940. https://doi.org/10.1016/j.jmb.2007.08.005
Pande J, Szewczyk MM, Grover AK (2010) Phage display: concept, innovations, applications and future. Biotechnol Adv 28:849–858. https://doi.org/10.1016/j.biotechadv.2010.07.004
Zahavi D, Weiner L (2020) Monoclonal antibodies in cancer therapy. Antibodies 9(349):34. https://doi.org/10.3390/ANTIB9030034
Alfaleh MA, Alsaab HO, Mahmoud AB et al (2020) Phage display derived monoclonal antibodies: from bench to bedside. Front Immunol 11:567223. https://doi.org/10.3389/FIMMU.2020.01986/BIBTEX
Metzker ML (2009) Sequencing technologies — the next generation. Nat Rev Genet 2010 111 11:31–46. https://doi.org/10.1038/nrg2626
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: 10 years of next-generation sequencing technologies. Nat Rev Genet 176(17):333–351. https://doi.org/10.1038/nrg.2016.49
Zhang J, Chiodini R, Badr A, Zhang G (2011) The impact of next-generation sequencing on genomics. J Genet Genomics 38:95–109. https://doi.org/10.1016/J.JGG.2011.02.003
Mardis ER (2013) Next-generation sequencing platforms. Annu Rev Anal Chem 6:287–303. https://doi.org/10.1146/annurev-anchem-062012-092628
Shendure J, Aiden EL (2012) The expanding scope of DNA sequencing. Nat Biotechnol 3011(30):1084–1094. https://doi.org/10.1038/nbt.2421
Meng W, Zhang B, Schwartz GW et al (2017) An atlas of B-cell clonal distribution in the human body. Nat Biotechnol 359(35):879–884. https://doi.org/10.1038/nbt.3942
Georgiou G, Ippolito GC, Beausang J et al (2014) The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotechnol 322(32):158–168. https://doi.org/10.1038/nbt.2782
Robins H (2013) Immunosequencing: applications of immune repertoire deep sequencing. Curr Opin Immunol 25:646–652. https://doi.org/10.1016/J.COI.2013.09.017
Friedensohn S, Neumeier D, Khan TA et al (2020) Convergent selection in antibody repertoires is revealed by deep learning. bioRxiv. https://doi.org/10.1101/2020.02.25.965673
Lim YW, Adler AS, Johnson DS (2022) Predicting antibody binders and generating synthetic antibodies using deep learning. MAbs. https://doi.org/10.1080/19420862.2022.2069075
Lecun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/NATURE14539
Sebastiani M, Vacchi C, Manfredi A, Cassone G (2022) Personalized medicine and machine learning: a roadmap for the future. J Clin Med. https://doi.org/10.3390/JCM11144110
Vadapalli S, Abdelhalim H, Zeeshan S, Ahmed Z (2022) Artificial intelligence and machine learning approaches using gene expression and variant data for personalized medicine. Brief Bioinform. https://doi.org/10.1093/BIB/BBAC191
Alt FW, Blackwell TK, Yancopoulos GD (1987) Development of the primary antibody repertoire. Science (80− ) 238:1079–1087. https://doi.org/10.1126/science.3317825
Rajewsky K (1996) Clonal selection and learning in the antibody system. Nature 381:751–758. https://doi.org/10.1038/381751a0
Yaari G, Kleinstein SH (2015) (2015) Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med 71(7):1–14. https://doi.org/10.1186/S13073-015-0243-2
Calis JJA, Rosenberg BR (2014) Characterizing immune repertoires by high throughput sequencing: strategies and applications. Trends Immunol 35:581–590. https://doi.org/10.1016/J.IT.2014.09.004
Jiang N, He J, Weinstein JA et al (2013) Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med 5:171ra19. https://doi.org/10.1126/scitranslmed.3004794
Lavinder JJ, Wine Y, Giesecke C et al (2014) Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc Natl Acad Sci USA 111:2259–2264. https://doi.org/10.1073/pnas.1317793111
Laserson U, Vigneault F, Gadala-Maria D et al (2014) High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci USA 111:4928–4933. https://doi.org/10.1073/pnas.1323862111
Roskin KM, Simchoni N, Liu Y et al (2015) IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci Transl Med 7:302ra135. https://doi.org/10.1126/scitranslmed.aab1216
Palanichamy A, Apeltsin L, Kuo TC et al (2014) Immunoglobulin class-switched B cells form an active immune axis between CNS and periphery in multiple sclerosis. Sci Transl Med 6:248ra106. https://doi.org/10.1126/scitranslmed.3008930
Robinson WH (2014) Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery. Nat Rev Rheumatol 113(11):171–182. https://doi.org/10.1038/nrrheum.2014.220
Lee YN, Frugoni F, Dobbs K et al (2016) Characterization of T and B cell repertoire diversity in patients with RAG deficiency. Sci Immunol. https://doi.org/10.1126/sciimmunol.aah6109
Friedensohn S, Khan TA, Reddy ST (2017) Advanced methodologies in high-throughput sequencing of immune repertoires. Trends Biotechnol 35:203–214. https://doi.org/10.1016/j.tibtech.2016.09.010
Reddy ST, Ge X, Miklos AE et al (2010) Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat Biotechnol 28:965–969
Zhu J, Ofek G, Yang Y et al (2013) Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains. Proc Natl Acad Sci USA 110:6470–6475. https://doi.org/10.1073/PNAS.1219320110/SUPPL_FILE/PNAS.201219320SI.PDF
Doria-Rose NA, Schramm CA, Gorman J et al (2014) Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nat 5097498(509):55–62. https://doi.org/10.1038/nature13036
Williams LD, Ofek G, Schätzle S et al (2017) Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci Immunol. https://doi.org/10.1126/sciimmunol.aal2200
Montague Z, Lv H, Otwinowski J et al (2021) Dynamics of B cell repertoires and emergence of cross-reactive responses in patients with different severities of COVID-19. Cell Rep 35:109173. https://doi.org/10.1016/J.CELREP.2021.109173
Il KS, Noh J, Kim S et al (2021) Stereotypic neutralizing VHantibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci Transl Med 13:27. https://doi.org/10.1126/SCITRANSLMED.ABD6990/SUPPL_FILE/PAPV3.PDF
Bashford-Rogers RJM, Bergamaschi L, McKinney EF et al (2019) B cell receptor repertoire analysis in six immune-mediated diseases. Nature 574:122. https://doi.org/10.1038/S41586-019-1595-3
Papalexi E, Satija R (2017) Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 181(18):35–45. https://doi.org/10.1038/nri.2017.76
Asensio MA, Lim YW, Wayham N et al (2019) Antibody repertoire analysis of mouse immunization protocols using microfluidics and molecular genomics. MAbs 11:870. https://doi.org/10.1080/19420862.2019.1583995
Shlesinger D, Hong KL, Shammas G et al (2022) Single-cell immune repertoire sequencing of B and T cells in murine models of infection and autoimmunity. Genes Immun 236(23):183–195. https://doi.org/10.1038/s41435-022-00180-w
Csepregi L, Ehling RA, Wagner B, Reddy ST (2020) Immune literacy: reading, writing, and editing adaptive immunity. iScience. https://doi.org/10.1016/J.ISCI.2020.101519
Chen H, Ye F, Guo G (2019) Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol 163(16):242–249. https://doi.org/10.1038/s41423-019-0214-4
García-Valiente R, Merino Tejero E, Stratigopoulou M et al (2023) Understanding repertoire sequencing data through a multiscale computational model of the germinal center. npj Syst Biol Appl. https://doi.org/10.1038/s41540-023-00271-y
Ostrovsky-Berman M, Frankel B, Polak P, Yaari G (2021) Immune2vec: embedding B/T cell receptor sequences in ℝ N using natural language processing. Front Immunol 12:6080687. https://doi.org/10.3389/fimmu.2021.680687
Kim I, Byun SY, Kim S et al (2021) Computational analysis of B cell receptor repertoires in COVID-19 patients using deep embedded representations of protein sequences. bioRxiv. https://doi.org/10.1101/2021.08.02.454701
Shiroguchi K, Jia TZ, Sims PA, Xie XS (2012) Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci USA 109:1347–1352. https://doi.org/10.1073/PNAS.1118018109/SUPPL_FILE/SD07.XLS
Turchaninova MA, Davydov A, Britanova OV et al (2016) High-quality full-length immunoglobulin profiling with unique molecular barcoding. Nat Protoc 119(11):1599–1616. https://doi.org/10.1038/nprot.2016.093
Khan TA, Friedensohn S, Gorter de Vries AR et al (2016) Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci Adv 2:e1501371. https://doi.org/10.1126/sciadv.1501371
Shugay M, Britanova OV, Merzlyak EM et al (2014) Towards error-free profiling of immune repertoires. Nat Methods 116(11):653–655. https://doi.org/10.1038/nmeth.2960
Dekosky BJ, Ippolito GC, Deschner RP et al (2013) High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat Biotechnol 312(31):166–169. https://doi.org/10.1038/nbt.2492
Goldstein LD, Chen YJJ, Wu J et al (2019) Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Commun Biol 21(2):1–10. https://doi.org/10.1038/s42003-019-0551-y
Friedensohn S, Lindner JM, Cornacchione V et al (2018) Synthetic standards combined with error and bias correction improve the accuracy and quantitative resolution of antibody repertoire sequencing in human naïve and memory B cells. Front Immunol 9:375992. https://doi.org/10.3389/FIMMU.2018.01401/BIBTEX
Acknowledgements
The author would like to thank Peter Olson and Janet Stewart for reviewing of this article and providing detailed suggestions.
Funding
This work was partially supported by RevivAb Educational Advancement Grant MT-021.
Author information
Authors and Affiliations
Contributions
EG made substantial contributions to the conception or design of the work; or the acquisition, analysis, or interpretation of data; or the creation of new software used in the work; EG drafted the work or revised it critically for important intellectual content; EG approved the version to be published; and EG agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gallo, E. Current advancements in B-cell receptor sequencing fast-track the development of synthetic antibodies. Mol Biol Rep 51, 134 (2024). https://doi.org/10.1007/s11033-023-08941-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-08941-0