Abstract
Background
Precise quantification of grafted human cells in preclinical animal models such as non-human primates, rodents and rabbits is needed for the evaluations of the safety and efficacy of cell therapy. Quantitative PCR (qPCR) as a swift, sensitive and powerful assay is suitable for human cell quantification. However, it is a formidable challenge due to that the genome of non-human primates share more than 95% of similarity as human. Methods: In the present study, we developed a probe-based quantitative PCR (qPCR) assay for the quantification of human cells in preclinical animal models via targeting human specific DNA in the intron of BRCA1 (termed BRCA1-qPCR). The 5′ and 3′ end of BRCA1-qPCR probe was conjugated with FAM and non-fluorescent quencher-minor groove binder (NFQ-MGB), respectively. 1 µg of genomic DNA from human and preclinical animal models including rhesus monkeys, cynomolgus monkeys, New Zealand white rabbits, SD rats, C57BL/6 and BALB/c mice were used for determining the specificity and sensitivity of the BRCA1-qPCR assay. A calibration curve was generated by BRCA1-qPCR analysis of linearized plasmid containing targeted human specific DNA in BRCA1. The BRCA1-qPCR assay was validated by analysis of 0.003%, 0.03% and 0.3% of human leukocytes mixed within murine leukocytes.
Results
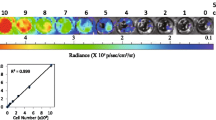
The BRCA1-qPCR assay detected human DNA rather than DNA from tested species. The amplification efficiency of the BRCA1-qPCR assay was 95.4% and the linearity of the calibration curve was R2 = 0.9997. The BRCA1-qPCR assay detected as low as 5 copies of human specific DNA and is efficient to specially amplify 30 pg human DNA in the presence of 1 µg of genomic DNA from tested species, respectively. The BRCA1-qPCR assay was able to quantify as low as 0.003% of human cells within murine leukocytes.
Conclusion
The BRCA1-qPCR assay is efficient for the quantification of human cells in preclinical animal models.




Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article.
Abbreviations
- qPCR:
-
quantitative PCR
- BLAST:
-
Basic local alignment search tool
References
El-Kadiry AE, Rafei M, Shammaa R (2021) Cell therapy: types, regulation, and clinical benefits. Front Med (Lausanne) 8:756029. https://doi.org/10.3389/fmed.2021.756029
Bastien JP, Minguy A, Dave V, Roy DC (2019) Cellular therapy approaches harnessing the power of the immune system for personalized cancer treatment. Semin Immunol 42:101306. https://doi.org/10.1016/j.smim.2019.101306
Wang LL et al (2021) Cell therapies in the clinic. Bioeng Transl Med 6:e10214. https://doi.org/10.1002/btm2.10214
Campbell A et al (2015) Concise review: process development considerations for cell therapy. Stem Cells Transl Med 4:1155–1163. https://doi.org/10.5966/sctm.2014-0294
Osada N (2015) Genetic diversity in humans and non-human primates and its evolutionary consequences. Genes Genet Syst 90:133–145. https://doi.org/10.1266/ggs.90.133
Harding J, Roberts RM, Mirochnitchenko O (2013) Large animal models for stem cell therapy. Stem Cell Res Ther 4:23. https://doi.org/10.1186/scrt171
Prior H et al (2020) Justification for species selection for pharmaceutical toxicity studies. Toxicol Res (Camb) 9:758–770. https://doi.org/10.1093/toxres/tfaa081
Song P et al (2012) Human genome-specific real-time PCR method for sensitive detection and reproducible quantitation of human cells in mice. Stem Cell Rev Rep 8:1155–1162. https://doi.org/10.1007/s12015-012-9406-3
Kline MC, Romsos EL, Duewer DL (2016) Evaluating digital PCR for the quantification of human genomic DNA: accessible amplifiable targets. Anal Chem 88:2132–2139. https://doi.org/10.1021/acs.analchem.5b03692
Yamamoto S, Ding N, Matsumoto SI, Hirabayashi H (2021) Highly specific, quantitative polymerase chain reaction probe for the quantification of human cells in cynomolgus monkeys. Drug Metab Pharmacokinet 36:100359. https://doi.org/10.1016/j.dmpk.2020.09.004
Suntsova MV, Buzdin AA (2020) Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genomics 21:535. https://doi.org/10.1186/s12864-020-06962-8
Lander ES et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. https://doi.org/10.1038/35057062
Lou DI et al (2014) Rapid evolution of BRCA1 and BRCA2 in humans and other primates. BMC Evol Biol 14:155. https://doi.org/10.1186/1471-2148-14-155
Somasundaram K (2003) Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair–perhaps through transcription. J Cell Biochem 88:1084–1091. https://doi.org/10.1002/jcb.10469
Lodovichi S et al (2020) Effect of BRCA1 missense variants on gene reversion in DNA double-strand break repair mutants and cell cycle-arrested cells of Saccharomyces cerevisiae. Mutagenesis 35:189–195. https://doi.org/10.1093/mutage/gez043
Huttley GA et al (2000) Adaptive evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Australian breast cancer family study. Nat Genet 25:410–413. https://doi.org/10.1038/78092
Fleming MA, Potter JD, Ramirez CJ, Ostrander GK, Ostrander EA (2003) Understanding missense mutations in the BRCA1 gene: an evolutionary approach. Proc Natl Acad Sci U S A 100:1151–1156. https://doi.org/10.1073/pnas.0237285100
Burk-Herrick A, Scally M, Amrine-Madsen H, Stanhope MJ, Springer MS (2006) Natural selection and mammalian BRCA1 sequences: elucidating functionally important sites relevant to breast cancer susceptibility in humans. Mamm Genome 17:257–270. https://doi.org/10.1007/s00335-005-0067-2
Kent WJ (2002) BLAT–the BLAST-like alignment tool. Genome Res 12:656–664. https://doi.org/10.1101/gr.229202
Soejima M, Hiroshige K, Yoshimoto J, Koda Y (2012) Selective quantification of human DNA by real-time PCR of FOXP2. Forensic Sci Int Genet 6:447–451. https://doi.org/10.1016/j.fsigen.2011.09.006
Zong C, Lu S, Chapman AR, Xie XS (2012) Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science 338:1622–1626. https://doi.org/10.1126/science.1229164
Hiroshige K, Soejima M, Nishioka T, Kamimura S, Koda Y (2009) Simple and sensitive method for identification of human DNA by allele-specific polymerase chain reaction of FOXP2. J Forensic Sci 54:857–861. https://doi.org/10.1111/j.1556-4029.2009.01063.x
Atkins JT et al (2020) Pre-clinical animal models are poor predictors of human toxicities in phase 1 oncology clinical trials. Br J Cancer 123:1496–1501. https://doi.org/10.1038/s41416-020-01033-x
Van Norman GA (2019) Limitations of animal studies for predicting toxicity in clinical trials: is it time to rethink our current approach? JACC Basic Transl Sci 4:845–854. https://doi.org/10.1016/j.jacbts.2019.10.008
Chua D, Low ZS, Cheam GX, Ng AS, Tan NS (2022) Utility of human relevant preclinical animal models in navigating NAFLD to MAFLD paradigm. Int J Mol Sci. https://doi.org/10.3390/ijms232314762
Shigeto J et al (2018) Preclinical toxicity studies for regenerative medicine in Japan. Clin Ther 40:1813–1822. https://doi.org/10.1016/j.clinthera.2018.09.007
Walker JA et al (2003) Human DNA quantitation using Alu element-based polymerase chain reaction. Anal Biochem 315:122–128. https://doi.org/10.1016/s0003-2697(03)00081-2
Walker JA et al (2005) Multiplex polymerase chain reaction for simultaneous quantitation of human nuclear, mitochondrial, and male Y-chromosome DNA: application in human identification. Anal Biochem 337:89–97. https://doi.org/10.1016/j.ab.2004.09.036
Funakoshi K et al (2017) Highly sensitive and specific Alu-based quantification of human cells among rodent cells. Sci Rep 7:13202. https://doi.org/10.1038/s41598-017-13402-3
Prigent J et al (2015) Human progenitor cell quantification after xenotransplantation in rat and mouse models by a sensitive qPCR assay. Cell Transpl 24:1639–1652. https://doi.org/10.3727/096368914X681955
Zhu K et al (2018) Lack of remuscularization following transplantation of human embryonic stem cell-derived cardiovascular progenitor cells in infarcted nonhuman primates. Circ Res 122:958–969. https://doi.org/10.1161/CIRCRESAHA.117.311578
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 82171791) and the Youth Innovation Team Grant and the Starting Grant by Xuzhou Medical University (grant no. D2018009), Xuzhou Technology Program (KC20089), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province, China (KYCX21_2638).
Author information
Authors and Affiliations
Contributions
KL, LH, SW, XC and YL performed the experiments. LH and KL performed data analysis. SZ collected human samples. LH, LL and HW conceived and supervised the study. LH prepared the figures and wrote the manuscript with input from HW and LL.
Corresponding authors
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
All animal studies were performed in accordance with the protocol approved by the Animal Experimental Ethics Committee of Xuzhou Medical University(Approval ID: 202211S011). The usage of blood samples was approved by the ethics committee of Nanjing First Hospital (Approval ID: KY20220516-04).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2023_8853_MOESM1_ESM.png
Figure S1. Analysis of human genomic DNA by the BRCA1-qPCR assay. The representative amplification plot of the BRCA1-qPCRanalysis of genomic DNA (1-10 ng) of 48 individuals (PNG 161.1 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, K., Hu, L., Wang, S. et al. An efficient qPCR assay for the quantification of human cells in preclinical animal models by targeting human specific DNA in the intron of BRCA1. Mol Biol Rep 50, 9229–9237 (2023). https://doi.org/10.1007/s11033-023-08853-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08853-z