Abstract
Background
Coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It is known that host microRNAs (miRNAs) can be modulated to favor viral infection or to protect the host. Herein, we report preliminary results of a study aiming at identifying differentially expressed plasmatic miRNAs in Brazilian patients with COVID-19.
Methods and results
miRNAs were extracted from the plasma of eight patients with COVID-19 (four patients with mild COVID-19 and four patients with severe/critical COVID-19) and four healthy controls. Patients and controls were matched for sex and age. miRNA expression levels were detected using high-throughput sequencing. Differential miRNA expression and enrichment analyses were further evaluated. A total of 18 miRNAs were differentially expressed between patients with COVID-19 and controls. miR-4433b-5p, miR-6780b-3p, miR-6883-3p, miR-320b, miR-7111-3p, miR-4755-3p, miR-320c, and miR-6511a-3p were the most important miRNAs significantly involved in the PI3K/AKT, Wnt/β-catenin, and STAT3 signaling pathways. Moreover, 42 miRNAs were differentially expressed between severe/critical and mild patients with COVID-19. miR-451a, miR-101-3p, miR-185-5p, miR-30d-5p, miR-25-3p, miR-342-3p, miR-30e-5p, miR-150-5p, miR-15b-5p, and miR-29c-3p were the most important miRNAs significantly involved in the Wnt/β-catenin, NF-κβ, and STAT3 signaling pathways.
Conclusions
If validated by quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) in a larger number of participants, the miRNAs identified in this study might be used as possible biomarkers for the diagnosis and severity of COVID-19.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease 2019 (COVID-19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), whose initial outbreak occurred in Wuhan, China. Unfortunately, this virus has spread rapidly worldwide, culminating in the pandemic situation we have been experiencing since mid-March 2020 [1]. SARS-CoV-2 is an RNA virus, which belongs to the genus Betacoronavirus, and is transmitted by inhalation of droplets containing the virus or by hand contact on surfaces containing the virus and subsequent contamination through the hands eyes, mouth, and nose. Symptoms include fever and dry cough in most patients, which may be accompanied by breathing difficulties, loss of taste, nausea, and diarrhea [2]. Severe cases of COVID-19 can progress to severe acute respiratory syndrome (SARS) and to a cytokine storm, in which the virus induces an intense inflammatory cascade [3]. Currently, there is no specific treatment for the management of COVID-19 [4]. Although some vaccines have been approved for emergency use by the Food and Drug Administration (FDA), such as the Comirnaty and Pfizer-BioNTech, Moderna, and Janssen (Johnson & Johnson) COVID-19 vaccines [5], and vaccine campaigns are ongoing, the emergence of new variants is a remaining concern [6].
Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) performed to detect viral RNA on nasopharyngeal or oropharyngeal swabs is the gold standard for the diagnosis of SARS-CoV-2. Although it is a specific and sensitive quantitative assay, there are some limitations, such as being relatively invasive and associated with an increased risk of cross-infection [7, 8]. Other methods, such as virus antigen or serological antibody testing, may also be used to diagnose COVID-19. While rapid antigen tests detect SARS-CoV-2 in samples from the respiratory tract, rapid serological tests detect antibodies (IgM and/or IgG) produced over days to weeks after infection by the virus. Both tests are easy to handle and require minimal personnel training. Rapid antibody tests, while less specific, can be useful for reducing exposure to risk factors during repeated sampling and are more cost effective compared to RT-PCR tests [7, 9]. Computed tomography of the chest consists of a complementary examination for the diagnosis of COVID-19, which allows the monitoring of disease evolution [7, 10]. In addition, the neutrophil–lymphocyte ratio (NLR) has been suggested as a prognostic marker for screening patients with COVID-19, with a higher NLR being associated with a poor prognosis [11]. Moreover, the detection of microRNAs (miRNAs) in human samples may be an alternative to identify effective biomarkers for the diagnosis and severity of COVID-19.
miRNAs are defined as small, single-stranded, non-coding RNA molecules of 21–23 nucleotides in length that bind to the target transcript in the 3' untranslated region (UTR), inhibiting protein translation and destabilizing their target messenger RNAs (mRNAs). miRNAs can regulate almost a third of the human genome and are widely involved in multiple pathways, such as cell proliferation, cell death, stress resistance, and fat metabolism. In addition, evidence suggests that a gain or loss of function of one or more miRNAs is associated with the diagnosis, progression, and prognosis of several cancers and infectious diseases [12, 13]. Therefore, dysregulated miRNAs, in addition to serving as biomarkers of disease, may be potential therapeutic targets providing a better understanding of the signaling pathways involved and the disease pathogenesis.
In a systematic review focused on the description of miRNAs associated with SARS-CoV-2 infection in human cells, only 2 out of the 29 studies included in the review reported the analysis of miRNA expression in patients with COVID-19 compared to non-COVID-19 samples [14]. Most of the studies included in this review reported miRNA data based on genome sequencing of SARS-CoV-2 isolates and computational approaches [14]. In addition, our research group conducted a scoping review on miRNAs differentially expressed in SARS-CoV-2 infected animals and patients with COVID-19, excluding studies based only on in silico prediction analysis [15]. Twenty studies were included, and 15 of which were conducted in patients. We have verified that miR-21-5p, miR-146a-5p, miR-126-3p, miR-144, and miR-155 may serve as important biomarkers of COVID-19 [15]. However, few studies have included participants from North and South America [15], where COVID-19 cases are rising again. Thus, the present article reports preliminary results (high-throughput sequencing analysis) of a study aiming at identifying differentially expressed plasmatic miRNAs in Brazilian patients with COVID-19 as possible biomarkers for disease diagnosis and severity.
Patients & methods
Study approval
The study was approved by the Research Ethics Committee of the School of Medical Sciences of the University of Campinas (UNICAMP) (numbers 36041420.0.000.5404 and 31049320.7.1001.5404). All participants or their guardians signed a consent form authorizing the use of their samples and data.
Participants and eligibility criteria
From May to September 2020, 12 participants were included in this study: four patients with severe/critical COVID-19 had been admitted to the Hospital Estadual Sumaré Dr. Leandro Francheschini (HES) in Sumaré city (SP, Brazil), affiliated with UNICAMP; four patients with mild COVID-19 had been admitted to the Hospital Municipal de Paulínia, in Paulínia city (SP, Brazil), and four healthy volunteers (controls).
The eligibility criteria were age ≥ 18 years and admission to HES or Hospital Municipal de Paulínia with a positive result in SARS-CoV-2 nasopharyngeal swab RT-PCR for patients with COVID-19, and age ≥ 18 years and negative result in SARS-CoV-2 nasopharyngeal swab RT-PCR for controls. Patients were excluded if their plasma samples were not sufficient to perform the experiments.
It is important to note that the control participants were not experiencing flu-like symptoms associated with COVID-19, had no contact with people infected with the SARS-CoV-2, were not front-line health professionals, and were followed up for 15 days after the collection of biological samples to ensure that they would not show COVID-19 related symptoms.
Demographic and clinical data
Data regarding the characteristics of patients with COVID-19 were obtained from medical records, including information concerning gender, age, race, and comorbidities. Additionally, they were classified by COVID-19 severity based on Falavigna et al.[16]: mild, presence of any signs and symptoms of COVID-19, but without dyspnea or abnormal chest imaging; moderate, evidence of disease in the lower respiratory tract and SpO2 > 93% on room air; severe, presence of one of the following factors: respiratory rate > 30 bpm, SpO2 ≤ 93% on room air, PaO2/FiO2 < 300 mm Hg, pulmonary infiltrate > 50%; and critical, respiratory failure, septic shock, and/or multiple organ dysfunction. Patients with severe/critical COVID-19 were also characterized by the length of hospital stay, drugs used for COVID-19 during hospitalization, the time they remained mechanically ventilated, prone position, and death. Data regarding control characteristics were obtained from interviews with volunteers, including information concerning age, gender, race, and comorbidities. Patients with severe/critical COVID-19, patients with mild COVID-19, and controls were matched for gender and age.
Sample collection and miRNA extraction
Venous whole blood samples were collected in EDTA-containing tubes from all participants (patients and controls). All patient samples were collected within ten days of the onset of the COVID-19 symptoms. Plasma samples were separated from whole blood by centrifugation at 2500 rpm, 4 °C for 10 min, and stored in freezer at − 80 °C until the experiments were carried out.
miRNA extraction was performed with 200 µL of each plasma sample using the miRNeasy Serum/Plasma Kit (Qiagen, Cat No. 217184), following the manufacturer’s instructions. At the end of the experiment, the samples were stored in a freezer at − 80 °C until further use.
Library construction and sequencing
Library construction was performed with 5 µL of each miRNA sample using the QIAseq® miRNA Library Kit (Qiagen, Cat No. 331502), following the manufacturer’s instructions. For quality control of the samples, 1 μL and 2 μL of each miRNA sequencing library was analyzed using an Agilent Bioanalyzer and a Qubit fluorometer, respectively, according to the manufacturer’s instructions. A quality check of the raw data is provided in Supplementary Table 1.
The samples were sent to the Life Sciences Core Facility (LaCTAD) from UNICAMP for sequencing. The library preparations were sequenced on an Illumina HiSeq 2500 platform, and 75 bp single-end reads were generated.
Bioinformatics analysis
Primary and secondary analyses were conducted in GeneGlobe. The primary analysis is based on counting the unique molecular identifiers (UMIs) and mapping the miRNA sequences, while the secondary analysis, using the UMI counts for each miRNA, performs the differential expression analysis. MiRWalk software was used to predict the miRNA target genes. MiRWalk provides predicted targets according to 12 different databases, including TargetScan [17]. A matrix was constructed to identify the interaction between miRNAs and their predicted target genes, which were sorted according to the potential target genes of different miRNAs. For diagnostic analysis, genes related to five or more miRNAs were selected for enrichment analysis, using the Ingenuity Pathway Analysis (IPA®, Qiagen) software to identify the main canonical signaling pathways involving differentially expressed miRNAs. For severity analysis, only target genes predicted by TargetScan and at least five different databases were selected for the following analyses. The genes targeted by at least two different miRNAs were selected for unsupervised enrichment analysis using the IPA software.
Statistical analysis
For the secondary analyses, the results were normalized using the DESeq2 method and the p values listed were returned by the Bioconductor software packages, such that p values less than 0.05, were considered significant. The results were expressed as fold-change (FC) and fold-regulation (FR). FC is the normalized miRNA expression in each test sample divided by the normalized miRNA expression in the control sample. FR represents the FC results in a biologically meaningful way. FC values greater than one indicate upregulation, and FR is equal to FC. FC values less than one indicate downregulation, and FR is the negative inverse of the FC. FR ≥ 1.6, or FR ≤ − 1.6, were used as criteria to select differentially expressed miRNAs for the diagnosis of COVID-19. FR ≥ 2.0, or FR ≤ − 2.0, were used as criteria to select miRNAs differentially expressed for COVID-19 severity.
For enrichment analysis (the most important canonical signaling pathways) of the predicted target genes, p values were calculated by Fisher’s exact test, and p values less than 0.05 were considered significant.
The Student’s t test (Microsoft Excel) was used to compare the mean age of the groups and a p value less than 0.05 was considered the cut-off for statistical significance.
Results
Characteristics of participants
Twelve participants were included in this study: patients with mild COVID-19 (mild, n = 4; moderate, n = 0; total, n = 4; sex: 2 male/2 female; mean age ± standard deviation [SD] 61.8 ± 11.7 years), patients with severe/critical COVID-19 [severe, n = 2; critical, n = 2; total, n = 4; sex: 2 male/2 female (severe: 1 male/1 female; critical: 1 male/1 female); mean age ± SD: 64.0 ± 8.6 years], and controls (n = 4; sex: 2 male/2 female; mean age ± SD: 62.8 ± 14.9 years). Sex and age were similar among the three groups (age comparison: controls vs. patients with COVID-19, p-value = 0.99; controls vs. mild COVID-19, p-value = 0.92; controls vs. severe/critical COVID-19, p value = 0.87; severe/critical vs. mild COVID-19, p value = 0.77). Table 1 shows the detailed characteristics of the participants.
Regarding patients with severe/critical COVID-19, the average length of stay was 36 ± 27 days; all patients received amoxicillin + clavulanate (1 g IV t.i.d.) and azithromycin (500 mg PO q.d.), two patients also received oseltamivir (75 mg PO b.i.d.) as antimicrobial treatment, two patients used dexamethasone (6 mg q.d.) to treat inflammation, and all patients were treated with anticoagulants (enoxaparin 40–60 mg/d SC or heparin 10,000–15,000 UI/d SC); they remained mechanically ventilated for 515 ± 346 h; two were pronated; and two patients died.
miRNAs as possible biomarkers of diagnosis for COVID-19
Differential miRNA expression profiling
A total of 18 miRNAs were differentially expressed between patients with COVID-19 and the controls, of which 13 were significantly upregulated and five were significantly downregulated (Fig. 1 and Table 2). The expression of miR-6780b-3p, miR-6883-3p, miR-4769-5p, miR-6873-3p, miR-320b, miR-7111-3p, miR-4755-3p, miR-320c, miR-6511a-3p, miR-320d, miR-5187-3p, miR-4508, and miR-4659a-5p was at least 1.6-fold higher in patients with COVID-19 than in controls. In addition, the expression of miR-4433b-5p, miR-16-2-3p, miR126-3p, miR-150-5p, and miR-224-5p was reduced by more than 1.6-fold in patients with COVID-19 compared with that in controls (Table 2).
Differential miRNA expression between patients with COVID-19 and controls (healthy volunteers). The abscissa presents the logarithmic value, logFC, of multiple differences in miRNA expression between the two groups and the ordinate represents the negative pair value of the p-value of the change in miRNA expression. Each point in the figure represents a miRNA. miRNAs with significant differences are represented by green (downregulated) and red (upregulated) dots. miRNAs without significant differences are represented by black dots
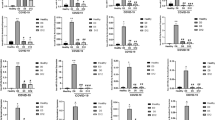
Additional analysis of only patients with mild COVID-19 compared to controls and only patients with severe/critical COVID-19 compared to controls were performed (Supplementary Table 2 and Supplementary Figs. 1 and 2). Only eight miRNAs were differentially expressed in the three analyses (Table 2 and Supplementary Table 2): miR-4433b-5p (downregulated), miR-6780b-3p, miR-6883-3p, miR-320b, miR-7111-3p, miR-4755-3p, miR-320c, and miR-6511a-3p (upregulated). Figure 2 shows a comparison of the expression levels of these miRNAs between the groups.
Expression of miRNAs in each sample. The abscissa presents the status of each participant: healthy volunteer (HV) (n = 4), patient with mild COVID-19 (M) (n = 4), patient with severe/critical COVID-19 (SC) (n = 4) and M + SC (n = 8). The letters b to h represent miRNAs consistently upregulated. The letter a represents the only miRNA consistently downregulated. Boxplot features: minimum whisker, the smallest value within; minimum box, 25th percentile; center, median; maximum box, 75th percentile; maximum whisker, the largest value within. ***p ≤ 0.001; **p ≤ 0.01; *p ≤ 0.05
Enrichment analysis
After the identification of the eight miRNAs cited above as possible diagnostic biomarkers, enrichment analysis was performed, and the top 50 canonical signaling pathways are shown in Fig. 3. Of the 50 most enriched signaling pathways, three appear to have an important role in modulating viral infections by miRNAs: Wnt/β-catenin signaling, phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) signaling, and the STAT3 pathway. A total of 242 genes were related to these pathways, and 64 of them were common to at least two of the above-mentioned pathways (data not shown). The matrix constructed for the analysis of miRNA-gene interactions indicated that miR-320b and miR-320c interacted the most with the 242 selected genes, and their interactions are described in more databases.
miRNAs as possible biomarkers of severity of COVID-19
Differential miRNA expression profiling
A total of 42 miRNAs were differentially expressed between severe/critical patients with COVID-19 and mild patients with COVID-19, all at least twofold lower in patients with severe/critical COVID-19 than in patients with mild COVID-19 (Fig. 4 and Table 3). miR-451a, miR-101-3p, miR-185-5p, miR-30d-5p, miR-25-3p, miR-342-3p, miR-30e-5p, miR-150-5p, miR-15b-5p, and miR-29c-3p were the 10 most downregulated miRNAs (Table 3).
Differential miRNA expression between patients with severe/critical and those with mild COVID-19. The abscissa presents the logarithmic value, logFC, of the multiple differences in miRNA expression between the two groups, and the ordinate presents the negative pair value of the p value of the change in miRNA expression. Each point in the figure represents a miRNA. Downregulated miRNAs with significant differences are represented by green dots. miRNAs without significant differences are represented by black dots. No miRNA is upregulated
As cited above, an analysis of patients with severe/critical COVID-19 compared to controls was also performed (Supplementary Table 2 and Supplementary Figs. 1 and 2).
Enrichment analysis
After the identification of 10 miRNAs cited as possible biomarkers of severity of COVID-19, enrichment analysis was performed, and the most important canonical signaling pathways are shown in Fig. 5. Three pathways, Wnt/β-catenin, NF-κβ, and STAT3 signaling, play an important role in viral infection and inflammatory response. A total of 159 genes were related to these pathways, and 34 of them were common to at least two of the above-mentioned pathways (data not shown). From the analysis of miRNA-gene interactions, miR-185-5p and miR-15b-5p are the ones that most interact with the 159 selected genes, and their interactions are described in more databases.
Discussion
miRNAs that enable the identification of SARS-CoV-2 infection may directly target the viral genome and proteins associated with the entry of the virus into the cell (e.g., ACE2, ADAM17, TMPRSS2), as well as regulate the immune system [18]. Based on this, a study suggested some miRNAs that could be important for COVID-19 diagnosis, such as miR-15b-5p, miR-195-5p, miR-221-3p, and miR-140-3p [18]. In our study, we identified eight important miRNAs as possible diagnostic biomarkers (miR-4433b-5p, miR-6780b-3p, miR-6883-3p, miR-320b, miR-7111-3p, miR-4755-3p, miR-320c, and miR-6511a-3p) that were not described in previously published articles on SARS-CoV-2 infected cells and animals, and patients with COVID-19 [15]. However, in a recent study, the most downregulated miRNA in our study, miR-4433b-5p, was differentially expressed between moderate and severe COVID-19 cases, suggesting that this miRNA might be a candidate for stratifying patients based on severity [19]. In our study, miR-4433b-5p was downregulated in both mild and severe/critical cases compared to controls; however, miR-4433b-5p was not differentially expressed between severe/critical cases and mild cases. In another recently published study, the miR-320 family, including miR-320b, was strongly downregulated in patients with COVID-19 induced severe respiratory failure compared to patients with COVID-19 induced moderate respiratory failure [20]. In our study, miR-320b was upregulated in both mild and severe/critical cases compared to controls; however, miR-320b was not differentially expressed between severe/critical cases and mild cases.
In addition, the expression of miR-16-2-3p was downregulated in patients with COVID-19 compared to controls, contrary to the studies by Li et al. in which the expression was upregulated [21, 22]. Moreover, miR-16-2-3p was found to be positively regulated in cells infected with SARS-CoV-2 compared to samples of normal human lung tissue [23]. High levels of this miRNA may play a role in mediating SARS-COV-2 infection [14]. However, the differential expression of miR-16-2-3p in patients with COVID-19 does not appear to be specific, since it was found in the comparison between patients with recent or complicated type 2 diabetes [24]. In addition, miR-16-2-3p expression was found to be upregulated in the plasma of patients with non-syndromic cleft lip [25] and downregulated in resistant glucocorticoid patients with ulcerative colitis [26].
We also found that miR-126-3p was differentially expressed and downregulated in patients with COVID-19 compared with that in controls, corroborating the findings of Sabbatinelli et al. [27] and Garg et al. [28]; however, these studies compared only patients with severe COVID-19 with healthy volunteers. Although we found miR-126-3p as a possible diagnostic biomarker in our study, miR-126-3p downregulation was associated with inflammation by regulating the NF-κβ inhibitor Iκ-Bα. Interestingly, miR-126-3p has also been associated with endothelial dysfunction, which can partially explain the vascular damage observed in the lungs of patients with COVID-19, associated with the presence of intracellular viruses and perivascular T-cell infiltrates [29], contributing to disease severity. Moreover, Mitchell et al. found miR-126-3p downregulated in severely ill COVID-19 hospitalized patients on mechanical ventilators when compared to mildly ill COVID-19 hospitalized patients [30].
In the comparison between patients with mild COVID-19 and controls, we found that miR-99a-5p and miR-378a-3p were upregulated (Supplementary Table 2). In a study by Tang et al. miR-99a-5p was found to be downregulated in severe cases compared to healthy volunteers and in severe cases compared to moderate cases and was associated with the expression of the proinflammatory genes IGF1R and MTMR3, which induce weaker antiviral immunity [31]. In addition, the increase in miR-378a-3p expression attenuated hypoxia-induced lesions in cardiomyocytes by suppressing the NEAT1 lncRNA [32], although in a study by Chen et al. elevated levels of miR-378a-3p were associated with a poor prognosis for COVID-19 [33].
In our study, from the eight most important miRNAs for the diagnosis of COVID-19, a total of 242 target genes were found to be possibly related to the Wnt/β-catenin signaling, PI3K/AKT signaling, and STAT3 pathways, which according to Barbu et al. are well known to be involved in viral infection. Wnt represents a group of highly preserved pathways in vertebrates through which extracellular signals are transported into the cell. In addition, there appears to be a relationship between the Wnt pathway and innate immune response in the host. However, there are theories that type I interferon (IFN) signaling can be increased while inhibiting the Wnt pathway [34]. STAT3 is a member of the STAT protein family that controls the expression of genes regulating different biological processes, such as immune responses, inflammation, and apoptosis. STAT3 protein is present in the cytoplasm in an inactive form and is activated by numerous types of cytokines, especially interleukin-6 (IL-6), which is a predictive marker of severity in COVID-19. Furthermore, STAT3 may have multiple roles during SARS-CoV-2 infection, including induction of pro-inflammatory responses, promotion of a cytokine storm, impairment of antiviral immune responses, alteration of virus replication, and exacerbation of lymphopenia [35]. Finally, with regard to the PI3K/AKT signaling pathway, it has an important role in viral replication, and its activation has been reported in many viruses, such as hepatitis B and C and human immunodeficiency. In addition, Shwetha et al. demonstrated that the upregulation of miR-320c during hepatitis C virus infection targets P13K/AKT signaling, which increases cell survival and corroborates the results found in our study [34, 36]. Therefore, these signaling pathways may be dysregulated in patients with COVID-19, thus contributing to its pathogenesis.
miRNAs as biomarkers of COVID-19 severity may be associated with the host immune response and inflammation and may serve as therapeutic targets [31]. Our study showed that miR-451a, miR-101-3p, miR-185-5p, miR-30d-5p, miR-25-3p, miR-342-3p, miR-30e-5p, miR-150-5p, miR-15b-5p, and miR-29c-3p were the ten most important downregulated miRNAs as possible biomarkers of disease severity. Among these miRNAs, miR-451a and miR-150-5p were found to be downregulated in plasma of patients with COVID-19 admitted in intensive care unit compared to those admitted to the ward in the study by Gonzalo-Calvo et al. and receiver operating characteristic (ROC) curve analysis showed that miR-451a was able to discriminate the COVID-19 severity with high accuracy [37]. Moreover, miR-15b-5p was found to be differentially expressed in two previous studies [18, 31]. According to Kim et al. miR-15b-5p directly binds to the viral genome, and in their study, it was also found to be downregulated during SARS-CoV-2 infection, which may allow the virus to escape the host immune defense by inhibiting apoptosis and promoting the proliferation of infected cells [18]. However, according to Tang et al. its upregulation can accelerate viral replication, mediate virus-induced changes in the cell transcriptome, and intensify the severity of COVID-19 [31]. miR-15b-5p was also predicted by three independent studies included in the systematic review published by Marchi et al. indicating that this miRNA can act by regulating host and SARS-CoV-2 genes [14].
The type I IFN signaling pathway is involved in viral infections [34]. Recent studies have shown that IFN is crucial for the severity of COVID-19, and its deficiency can lead to severe disease [38, 39]. However, the type I IFN pathway appears not to be significantly enriched in the ten downregulated miRNAs in severe/critical COVID-19 patients. On the other hand, our study showed that these downregulated miRNAs appear to be involved in the Wnt/β-catenin signaling pathway, an immune regulation pathway that may be hyperactivated. Once type I IFN signaling can be increased while inhibiting the Wnt pathway [34], it is possible that the hyperactivated Wnt signaling pathway is related to decreased type I IFN signaling pathway. Moreover, NF-κβ and STAT3 signaling pathways appear to be significantly enriched, and they are also known to be involved in viral infection [34]. It is known that the severity of COVID-19 is dependent on a cytokine storm, most likely induced by the IL-6 amplifier, which is a hyperactivation machinery that regulates the NF-κB pathway and is stimulated by the simultaneous activation of STAT3 and NF-κB signaling in non-immune cells, including alveolar epithelial cells and endothelial cells [40]. Therefore, the hyperactivation of these two signaling pathways plays a key role in the inflammation of non-immune cells and may cause fatal symptoms such as acute respiratory distress syndrome (ARDS), severe pneumonia, multiorgan failure, and coagulation [40]. The signaling pathways found may be possible therapeutic targets for the treatment of patients with severe or critical COVID-19.
Additional downregulated miRNAs identified in this study have been related to the disease severity, including miR-195-5p, miR-140-5p, miR-144-3p, miR-125a-5p, miR-30a-5p, let-7f-5p, let-7 g-5p, and let-7i-5p (Table 3; severe/critical compared to mild COVID-19), and miR-195-5p, miR-144-3p, miR-125a-5p, miR-30a-5p, miR-21-5p, let-7a-5p, let-7d-5p, and let-7f-5p (Supplementary Table 2; severe/critical COVID-19 compared to control). According to Kim et al. miR-195-5p directly binds to the viral genome, but unlike our results, this miRNA was upregulated in SARS-CoV-2 infection and appears to be related to the promotion of apoptosis by inducing cell cycle arrest and the prevention of excessive proliferation of infected cells [18]. Moreover, miR-140-3p and miR-144-3p were found to be downregulated in the studies by Kim et al. and Li et al. respectively, as well as in our work [18, 21, 22]; however, Li et al. [21, 22] studied miRNAs as diagnostic biomarkers (patients with mild or moderate COVID-19 compared to healthy volunteers). It is known that miR-140-3p targets TMPRSS2, and its downregulation can contribute to viral infection by inhibiting apoptosis and promoting cell proliferation [18]. The pathophysiological implication of miR-144-3p dysregulation has not been reported by Li et al. [21, 22]. In contrast, Guo et al. showed that this miRNA is upregulated in the lung tissues of mice infected with the H7N9/AH1-PB2-627E/701 N strain of the influenza virus [41]. Furthermore, downregulation of miR-125a-5p and miR-30 may favor entry of the virus into the cell. miR-125a-5p was predicted to directly target ACE2 mRNA or associated ACE2 pathways, while miR-30a was found to be negatively correlated with the TMPRSS2 protein [14]. According to Tang et al. and corroborating our results and those of Sabatinelli et al. the downregulation of miR-21-5p is associated with inflammation [27, 31], while its upregulation appears to be associated with cardiac fibrosis [28]. Finally, the altered expression of members of the let-7 family found in our work supports the findings of Chen et al. and Zheng et al. revealing the importance of this family in the activation of T cells and during the inflammatory response [33, 42]. Indeed, let-7 miRNAs play a key role in the activation of the immune system and the inflammatory response by targeting the IL-6 gene and reducing its expression [43, 44].
This study has some limitations: (a) the blood samples were collected at different times after the onset of COVID-19 symptoms, i.e., different periods of infection between patients; (b) we did not include non-COVID-19 patients with respiratory viral infection as positive controls (e.g., patients infected with influenza virus) to validate the specificity of these miRNAs as biomarkers of COVID-19; (c) we also did not include asymptomatic SARS-CoV-2 infected patients, and thus, it is not known if the differential expression of these miRNAs would be useful for diagnosing these patients; (d) we do not know which strains of SARS-CoV-2 infected the patients studied; (e) although we matched age and gender between healthy volunteers and patients with COVID-19, race and comorbidities, such as type 2 diabetes, obesity and coronary artery diseases, that are known to alter the expression of miRNAs could not be precisely combined in our study; and (f) a small number of participants were included in the present study; however, the next step of our research is to validate the main miRNAs herein identified by quantitative RT-PCR (preferred method for the identification of miRNAs) in a larger cohort. Our results can also guide future studies to be conducted by other research groups, which can be performed in patients of other ethnicities.
Conclusion
In conclusion, the differentially expressed miRNAs identified in this study might be used as possible biomarkers for the diagnosis and severity of COVID-19 after being validated by quantitative RT-PCR and in a larger number of participants. Moreover, these miRNAs and signaling pathways may be possible targets for developing therapeutics for treating COVID-19.
References
Lu H, Stratton CW, Tang YW (2020) Outbreak of pneumonia of unknown etiology in Wuhan, China: the mystery and the miracle. J Med Virol 92(4):401–402. https://doi.org/10.1002/jmv.25678
Guan WJ, Ni ZY, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382(18):1708–1720. https://doi.org/10.1056/NEJMoa2002032
Coperchini F, Chiovato L, Croce L et al (2020) The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 53:25–32. https://doi.org/10.1016/j.cytogfr.2020.05.003
Jean SS, Lee PI, Hsueh PR (2020) Treatment options for COVID-19: the reality and challenges. J Microbiol Immunol Infect 53(3):436–443. https://doi.org/10.1016/j.jmii.2020.03.034
FDA. Covid-19 vaccines authorized for emergency use or FDA-approved. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines. Accessed 15 September 2021
Chung JY, Thone MN, Kwon YJ (2021) COVID-19 vaccines: the status and perspectives in delivery points of view. Adv Drug Deliv Rev 170:1–25. https://doi.org/10.1016/j.addr.2020.12.011
Ji T, Liu Z, Wang G et al (2021) Detection of COVID-19: a review of the current literature and future perspectives. Biosens Bioelectron 166:112455. https://doi.org/10.1016/j.bios.2020.112455
Adeoye J, Thomson P (2020) ‘The double-edged sword’—an hypothesis for Covid-19-induced salivary biomarkers. Med Hypotheses 143:110124. https://doi.org/10.1016/j.mehy.2020.110124
Castro R, Luz PM, Wakimoto MD et al (2020) COVID-19: a meta-analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz J Infect Dis 24(2):180–187. https://doi.org/10.1016/j.bjid.2020.04.003
Jin YH, Zhan QY, Peng ZY et al (2020) Chemoprophylaxis, diagnosis, treatments, and discharge management of COVID-19: an evidence-based clinical practice guideline (updated version). Mil Med Res 7(1):41. https://doi.org/10.1186/s40779-020-00270-8
Pimentel GD, Dela Vega MCM, Laviano A (2020) High neutrophil to lymphocyte ratio as a prognostic marker in COVID-19 patients. Clin Nutr ESPEN 40:101–102. https://doi.org/10.1016/j.clnesp.2020.08.004
Mishra PJ, Bertino JR (2009) MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics 10(3):399–416. https://doi.org/10.2217/14622416.10.3.399
Correia CN, Nalpas NC, McLoughlin KE et al (2017) Circulating microRNAs as potential biomarkers of infectious disease. Front Immunol 8:118. https://doi.org/10.3389/fimmu.2017.00118
Marchi R, Sugita B, Centa A et al (2021) The role of microRNAs in modulating SARS-CoV-2 infection in human cells: a systematic review. Infect Genet Evol 91:104832. https://doi.org/10.1016/j.meegid.2021.104832
Visacri MB, Nicoletti AS, Pincinato EC et al (2021) MicroRNAs as biomarkers of COVID-19: a scoping review on role, status, and future directions for research in this field. Biomark Med 15(18):1785–1795. https://doi.org/10.2217/bmm-2021-0348
Falavigna M, Colpani V, Stein C et al (2020) Guidelines for the pharmacological treatment of COVID-19. The task-force/consensus guideline of the Brazilian association of intensive care medicine, the Brazilian society of infectious diseases and the Brazilian society of pulmonology and tisiology. Rev Bras Ter Intensiva 32(2):166–196. https://doi.org/10.5935/0103-507x.20200039
Agarwal V, Bell GW, Nam JW, Bartel DP (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife 4:e05005. https://doi.org/10.7554/eLife.05005
Kim WR, Park EG, Kang KW et al (2020) Expression analyses of microRNAs in hamster lung tissues infected by SARS-CoV-2. Mol Cells 43(11):953–963. https://doi.org/10.14348/molcells.2020.0177
Farr RJ, Rootes CL, Rowntree LC et al (2021) Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog 17(7):1009759. https://doi.org/10.1371/journal.ppat.1009759
Duecker RP, Adam EH, Wirtz S et al (2021) The MiR-320 family is strongly downregulated in patients with COVID-19 induced severe respiratory failure. Int J Mol Sci 22(19):10351. https://doi.org/10.3390/ijms221910351
Li C, Hu X, Li L, Li JH (2020) Differential microRNA expression in the peripheral blood from human patients with COVID-19. J Clin Lab Anal 34(10):23590. https://doi.org/10.1002/jcla.23590
Li CX, Chen J, Lv SK et al (2021) Whole-transcriptome RNA sequencing reveals significant differentially expressed mRNAs, miRNAs, and lncRNAs and related regulating biological pathways in the peripheral blood of COVID-19 patients. Mediators Inflamm 2021:6635925. https://doi.org/10.1155/2021/6635925
Chow JT, Salmena L (2020) Prediction and analysis of SARS-CoV-2-targeting microRNA in human lung epithelium. Genes (Basel) 11(9):1002. https://doi.org/10.3390/genes11091002
Meerson A, Najjar A, Saad E et al (2019) Sex differences in plasma microRNA biomarkers of early and complicated diabetes mellitus in israeli arab and jewish patients. Noncoding RNA 5(2):32. https://doi.org/10.3390/ncrna5020032
Zou J, Li J, Ji C et al (2016) Expression profile of plasma microRNAs in nonsyndromic cleft lip and their clinical significance as biomarkers. Biomed Pharmacother 82:459–466. https://doi.org/10.1016/j.biopha.2016.05.033
Luo J, Wang Y, Lan D et al (2018) Differential expression of serum microRNAs in glucocorticoid-resistant patients with ulcerative colitis. Int J Clin Exp Pathol 11(2):936–946
Sabbatinelli J, Giuliani A, Matacchione G et al (2021) Decreased serum levels of the inflammaging marker miR-146a are associated with clinical response to tocilizumab in COVID-19 patients. Mech Ageing Dev 193:111413. https://doi.org/10.1016/j.mad.2020.111413
Garg A, Seeliger B, Derda AA et al (2021) Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur J Heart Fail 23(3):468–475. https://doi.org/10.1002/ejhf.2096
Ackermann M, Verleden SE, Kuehnel M et al (2020) Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med 383(2):120–128. https://doi.org/10.1056/NEJMoa2015432
Mitchell MI, Ben-Dov IZ, Liu C et al (2021) Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): a customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J Extracell Vesicles 10(8):e12110. https://doi.org/10.1002/jev2.12110
Tang H, Gao Y, Li Z et al (2020) The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin Transl Med 10(6):e200. https://doi.org/10.1002/ctm2.200
Zhao J, Chen F, Ma W, Zhang P (2020) Suppression of long noncoding RNA NEAT1 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-378a-3p. Gene 731:144324. https://doi.org/10.1016/j.gene.2019.144324
Chen YM, Zheng Y, Yu Y et al (2020) Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J 39(24):e105896. https://doi.org/10.15252/embj.2020105896
Barbu MG, Condrat CE, Thompson DC et al (2020) MicroRNA involvement in signaling pathways during viral infection. Front Cell Dev Biol 8:143. https://doi.org/10.3389/fcell.2020.00143
Jafarzadeh A, Nemati M, Jafarzadeh S (2021) Contribution of STAT3 to the pathogenesis of COVID-19. Microb Pathog 154:104836. https://doi.org/10.1016/j.micpath.2021.104836
Shwetha S, Gouthamchandra K, Chandra M et al (2013) Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep 3:1555. https://doi.org/10.1038/srep01555
de Gonzalo-Calvo D, Benítez ID, Pinilla L et al (2021) Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl Res 236:147–159. https://doi.org/10.1016/j.trsl.2021.05.004
Hadjadj J, Yatim N, Barnabei L et al (2020) Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369(6504):718–724. https://doi.org/10.1126/science.abc6027
Meffre E, Iwasaki A (2020) Interferon deficiency can lead to severe COVID. Nature 587(7834):374–376. https://doi.org/10.1038/d41586-020-03070-1
Hojyo S, Uchida M, Tanaka K et al (2020) How COVID-19 induces cytokine storm with high mortality. Inflamm Regen 40:37. https://doi.org/10.1186/s41232-020-00146-3
Guo Y, Huang N, Tian M et al (2020) Integrated analysis of microRNA-mRNA expression in mouse lungs infected with H7N9 influenza virus: a direct comparison of host-adapting PB2 mutants. Front Microbiol 11:1762. https://doi.org/10.3389/fmicb.2020.01762
Zheng HY, Xu M, Yang CX et al (2020) Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct Target Ther 5(1):294. https://doi.org/10.1038/s41392-020-00457-4
Abu-Izneid T, AlHajri N, Mohammed Ibrahim A et al (2021) Micro-RNAs in the regulation of immune response against SARS COV-2 and other viral infections. J Adv Res 30:133–145. https://doi.org/10.1016/j.jare.2020.11.013
Sung SY, Liao CH, Wu HP et al (2013) Loss of let-7 microRNA upregulates IL-6 in bone marrow-derived mesenchymal stem cells triggering a reactive stromal response to prostate cancer. PLoS ONE 8(8):71637. https://doi.org/10.1371/journal.pone.0071637
Acknowledgements
We thank the staff of the Life Sciences Core Facility (LaCTAD) from University of Campinas (UNICAMP), for the quality control of the samples and sequencing.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) [Finance Code 001—88881.504454/2020-01 and 88887.506965/2020-00], by São Paulo Research Foundation (FAPESP) [number 2021/04669-9], and by the National Council for Scientific and Technological Development (CNPq). ASN is a recipient of a doctoral scholarship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) [number 88887.511334/2020-00]. MBV is a recipient of a postdoctoral scholarship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) [number 88887.504453/2020-00]. CRSCR is a recipient of a doctoral scholarship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) [number 88887.513100/2020-00].
Author information
Authors and Affiliations
Contributions
PM contributed with conception of the work. ASN, MBV, CRSCR, and RNS contributed to data acquisition. CRSCR, DSV, AE, LFSS, MWPJ, RRC, LOR, LAS, ND, and WJF contributed with the recruitment of the participants and obtaining plasma samples. MBV, ASN, PENSV, PM, and ECP performed the experiments (miRNA extraction and library construction). PM, MBV, ASN, PENSV, and JCFQ performed bioinformatics and statistical analysis. ML performed the SARS-CoV-2 nasopharyngeal swab RT-PCR for control participants. PM and JLC supervised the study. MBV and ASN wrote the manuscript. PM, ECP, and JCFQ revised the manuscript. All authors have read and agreed with the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Research Ethics Committee of the School of Medical Sciences of the University of Campinas (UNICAMP) [numbers 36041420.0.000.5404 and 31049320.7.1001.5404].
Consent for publication
A preprint of this article has been already submitted on ResearchSquare plataform (https://doi.org/10.21203/rs.3.rs-630726/v1) and published under a CC BY 4.0 License.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2022_7338_MOESM3_ESM.pdf
Supplementary file3 (PDF 41 KB) Supplementary Figure 1 Differential miRNA expression between patients with mild COVID-19 and controls (healthy volunteers). The abscissa presents the logarithmic value, logFC, of multiple differences in miRNA expression between the two groups and the ordinate represents the negative pair value of the p value of the change in miRNA expression. Each point in the figure represents a miRNA. miRNAs with significant differences are represented by blue (downregulated) and red (upregulated) dots. miRNAs without significant differences are represented by black dots
11033_2022_7338_MOESM4_ESM.pdf
Supplementary file4 (PDF 50 KB) Supplementary Figure 2 Differential miRNA expression between patients with severe/critical COVID-19 and controls (healthy volunteers). The abscissa presents the logarithmic value, logFC, of multiple differences in miRNA expression between the two groups and the ordinate represents the negative pair value of the p value of the change in miRNA expression. Each point in the figure represents a miRNA. miRNAs with significant differences are represented by blue (downregulated) and red (upregulated) dots. miRNAs without significant differences are represented by black dots
Rights and permissions
About this article
Cite this article
Nicoletti, A.d., Visacri, M.B., da Ronda, C.R.d. et al. Differentially expressed plasmatic microRNAs in Brazilian patients with Coronavirus disease 2019 (COVID-19): preliminary results. Mol Biol Rep 49, 6931–6943 (2022). https://doi.org/10.1007/s11033-022-07338-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07338-9