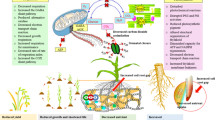
Abstract
An increase in ambient temperature throughout the twenty-first century has been described as a “worldwide threat” for crop production. Due to their sessile lifestyles, plants have evolved highly sophisticated and complex heat stress response (HSR) mechanisms to respond to higher temperatures. The HSR allows plants to minimize the damages caused by heat stress (HS), thus enabling cellular protection. HSR is crucial for their lifecycle and yield, particularly for plants grown in the field. At the cellular level, HSR involves the production of heat shock proteins (HSPs) and other stress-responsive proteins to counter the negative effects of HS. The expression of most HSPs is transcriptionally regulated by heat shock transcription factors (HSFs). HSFs are a group of evolutionary conserved regulatory proteins present in all eukaryotes and regulate various stress responses and biological processes in plants. In recent years, significant progress has been made in deciphering the complex regulatory network of HSFs, and several HSFs not only from model plants but also from major crops have been functionally characterized. Therefore, this review explores the progress made in this fascinating research area and debates the further potential to breed thermotolerant crop cultivars through the modulation of HSF networks. Furthermore, we discussed the role of HSFs in plant HS tolerance in a class-specific manner and shed light on their functional diversity, which is evident from their mode of action. Additionally, some research gaps have been highlighted concerning class-specific manners.
Similar content being viewed by others
Data availability
Not applicable.
Material availability
Not applicable.
Code availability
Not applicable.
Abbreviations
- HS:
-
Heat stress
- HSR:
-
Heat stress response
- HSFs:
-
Heat shock transcription factors
- HSPs:
-
Heat shock proteins
- TF:
-
Transcription factor
- ROS:
-
Reactive oxygen species
- DBD:
-
DNA-binding domain
- OD:
-
Oligomerization domain
- HR-A/B:
-
Hydrophobic associated-A/B region
- NLS:
-
Nuclear localization signal
- NES:
-
Nuclear export signal
- AHA:
-
Activator peptide motif
- HSEs:
-
Heat shock elements
- RD:
-
Repression domain
- HTH:
-
Helix-turn-helix motif
- OE:
-
Overexpression
- CS:
-
Co-suppression
- JA:
-
Jasmonic acid
- UPR:
-
Unfolded protein response
- APX2:
-
Ascorbate peroxidase 2
- H2O2 :
-
Hydrogen peroxide
- DREB2A:
-
Dehydration-responsive element-binding protein 2A
- OsGolS1:
-
Oryza sativa Galactinol synthase 1
- ABA:
-
Abscisic acid
- BRs:
-
Brassinosteroid
References
Hunter MC, Smith RG, Schipanski ME, Atwood LW, Mortensen DA (2017) Agriculture in 2050: recalibrating targets for sustainable intensification. Bioscience 67(4):386–391
Haider S, Iqbal J, Naseer S, Shaukat M, Abbasi BA, Yaseen T et al (2021) Unfolding molecular switches in plant heat stress resistance: a comprehensive review. Plant Cell Rep. https://doi.org/10.1007/s00299-021-02754-w
Raza A, Ashraf F, Zou X, Zhang X, Tosif H (2020) Plant adaptation and tolerance to environmental stresses: mechanisms and perspectives. Plant ecophysiology and adaptation under climate change: mechanisms and perspectives I. Springer, Singapore, pp 117–145
Zahra N, Shaukat K, Hafeez MB, Raza A, Hussain S, Chaudhary MT, Akram MZ, Kakavand SN, Saddiq MS, Wahid A (2021) Physiological and molecular responses to high, chilling, and freezing temperature in plant growth and production: consequences and mitigation possibilities. Harsh environment and plant resilience: molecular and functional aspects. New York, Springer
Stocker T (ed) (2014) Climate change 2013: the physical science basis: Working Group I contribution to the Fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge
Haider S, Iqbal J, Naseer S, Yaseen T, Shaukat M, Bibi H et al (2021) Molecular mechanisms of plant tolerance to heat stress: current landscape and future perspectives. Plant Cell Rep. https://doi.org/10.1007/s00299-021-02696-3
Hatfield JL, Prueger JH (2015) Temperature extremes: Effect on plant growth and development. Weather Clim Extrem 10:4–10
Janni M, Gullì M, Maestri E, Marmiroli M, Valliyodan B, Nguyen HT, Marmiroli N (2020) Molecular and genetic bases of heat stress responses in crop plants and breeding for increased resilience and productivity. J Exp Bot 71(13):3780–3802
Hussain S, Khaliq A, Ali B, Hussain HA, Qadir T, Hussain S (2019) Temperature extremes: impact on rice growth and development. Plant abiotic stress tolerance. Springer, Cham, pp 153–171
Raza A (2020) Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. https://doi.org/10.1007/s00299-020-02635-8
Zhao J, Lu Z, Wang L, Jin B (2021) Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. Int J Mol Sci 22(1):117
Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295(5561):1852–1858
Jacob P, Hirt H, Bendahmane A (2017) The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol J 15(4):405–414
Raza A, Tabassum J, Kudapa H, Varshney RK (2021) Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol 41:1209–1232
Haslbeck M, Vierling E (2015) A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol 427(7):1537–1548
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167(2):313–324
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22(1):53–65
Fragkostefanakis S, Röth S, Schleiff E, Scharf KD (2015) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38(9):1881–1895
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (HSF) family: structure, function and evolution. Biochim Biophys Acta (BBA) Gene Regul Mech 1819(2):104–119
Nover L, Scharf KD (1997) Heat stress proteins and transcription factors. Cell Mol Life Sci CMLS 53(1):80–103
Nover L, Scharf KD, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley WB (1996) The HSF world: classification and properties of plant heat stress transcription factors. Cell Stress Chaperones 1(4):215
Pirkkala L, Nykänen P, Sistonen LEA (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15(7):1118–1131
Voellmy R (2004) On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9(2):122
Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6(3):177
Haider S, Rehman S, Ahmad Y, Raza A, Tabassum J, Javed T, Osman HS, Mahmood T (2021) In silico characterization and expression profiles of heat shock transcription factors (HSFs) in Maize (Zea mays L.). Agronomy 11:2335. https://doi.org/10.3390/agronomy11112335
Ye J, Yang X, Hu G, Liu Q, Li W, Zhang L, Song X (2020) Genome-wide investigation of heat shock transcription factor family in wheat (Triticum aestivum L.) and possible roles in anther development. Int J Mol Sci 21(2):608
von Koskull-Döring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12(10):452–457
Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf KD (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10(3):310–316
Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH (2016) The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front Plant Sci 7:114
Kotak S, Vierling E, Bäumlein H, von Koskull-Döring P (2007) A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19(1):182–195
Harrison CJ, Bohm AA, Nelson HC (1994) Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263(5144):224–227
Peteranderl R, Rabenstein M, Shin YK, Liu CW, Wemmer DE, King DS, Nelson HC (1999) Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 38(12):3559–3569
Czarnecka-Verner E, Pan S, Salem T, Gurley WB (2004) Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol Biol 56(1):57–75
Andrási N, Pettkó-Szandtner A, Szabados L (2021) Diversity of plant heat shock factors: regulation, interactions, and functions. J Exp Bot 72(5):1558–1575
Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf KD (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16(12):1555–1567
Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K et al (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286(5):321–332
Liu HC, Liao HT, Charng YY (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant, Cell Environ 34(5):738–751
Liu HC, Charng YY (2013) Common and distinct functions of Arabidopsis class A1 and A2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol 163(1):276–290
Albihlal WS, Obomighie I, Blein T, Persad R, Chernukhin I, Crespi M et al (2018) Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b regulates multiple developmental genes under benign and stress conditions. J Exp Bot 69(11):2847–2862
Tian X, Wang F, Zhao Y, Lan T, Yu K, Zhang L et al (2020) Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol J 18(5):1109
Shah Z, Shah SH, Ali GS, Munir I, Khan RS, Iqbal A et al (2020) Introduction of Arabidopsis’s heat shock factor HSFA1d mitigates adverse effects of heat stress on potato (Solanum tuberosum L.) plant. Cell Stress Chaperones 25(1):57–63
Li G, Zhang Y, Zhang H, Zhang Y, Zhao L, Liu Z, Guo X (2019) Characteristics and regulating role in thermotolerance of the heat shock transcription factor ZmHSF12 from Zea mays L. J Plant Biol 62(5):329–341
Li HC, Zhang HN, Li GL, Liu ZH, Zhang YM, Zhang HM, Guo XL (2015) Expression of maize heat shock transcription factor gene ZmHsf06 enhances the thermotolerance and drought-stress tolerance of transgenic Arabidopsis. Funct Plant Biol 42(11):1080–1091
Gai WX, Ma X, Li Y, Xiao JJ, Khan A, Li QH, Gong ZH (2020) CaHsfA1d improves plant thermotolerance via regulating the expression of stress-and antioxidant-related genes. Int J Mol Sci 21(21):8374
Hafeez MB, Zahra N, Zahra K, Raza A, Khan A, Shaukat K, Khan S (2021) Brassinosteroids: molecular and physiological responses in plant growth and abiotic stresses. Plant Stress 2:100029
Nolan TM, Vukašinović N, Liu D, Russinova E, Yin Y (2020) Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32(2):295–318
Albertos P, Dündar G, Schenk P, Carrera S, Cavelius P, Sieberer T, Poppenberger B (2022) Transcription factor BES1 interacts with HSFA1 to promote heat stress resistance of plants. EMBO J. https://doi.org/10.15252/embj.2021108664
Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT (2007) A heat-inducible transcription factor, HSFA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143(1):251–262
Haider S, Iqbal J, Shaukat M, Naseer S, Mahmood T (2021) The epigenetic chromatin-based regulation of somatic heat stress memory in plants. Plant Gene 27:100318
Lämke J, Brzezinka K, Altmann S, Bäurle I (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J 35(2):162–175
Fragkostefanakis S, Mesihovic A, Simm S, Paupière MJ, Hu Y, Paul P et al (2016) HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol 170(4):2461–2477
Hu Y, Mesihovic A, Jiménez-Gómez JM, Röth S, Gebhardt P, Bublak D et al (2020) Natural variation in HsfA2 pre-mRNA splicing is associated with changes in thermotolerance during tomato domestication. New Phytol 225(3):1297–1310
Cheng Q, Zhou Y, Liu Z, Zhang L, Song G, Guo Z et al (2015) An alternatively spliced heat shock transcription factor, OsHSFA 2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol 17(2):419–429
Hue NT, Tran HT, Phan T, Nakamura J, Iwata T, Harano K et al (2013) Hsp90 and reactive oxygen species regulate thermotolerance of rice seedlings via induction of heat shock factor A2 (OsHSFA2) and galactinol synthase 1 (OsGolS1). Agric Sci 4:154–164
Guo XL, Yuan SN, Zhang HN, Zhang YY, Zhang YJ, Wang GY et al (2020) Heat-response patterns of the heat shock transcription factor family in advanced development stages of wheat (Triticum aestivum L.) and thermotolerance-regulation by TaHsfA2–10. BMC Plant Biol 20(1):1–18
Liu Z, Li G, Zhang H, Zhang Y, Zhang Y, Duan S et al (2020) TaHsfA2–1, a new gene for thermotolerance in wheat seedlings: characterization and functional roles. J Plant Physiol 246:153135
Lin KF, Tsai MY, Lu CA, Wu SJ, Yeh CH (2018) The roles of Arabidopsis HSFA2, HSFA4a, and HSFA7a in the heat shock response and cytosolic protein response. Bot Stud 59(1):1–9
Xin H, Zhang H, Zhong X, Lian Q, Dong A, Cao L et al (2017) Over-expression of LlHsfA2b, a lily heat shock transcription factor lacking trans-activation activity in yeast, can enhance tolerance to heat and oxidative stress in transgenic Arabidopsis seedlings. Plant Cell Tissue Organ Cult PCTOC 130(3):617–629
Zhang H, Li G, Hu D, Zhang Y, Zhang Y, Shao H et al (2020) Functional characterization of maize heat shock transcription factor gene ZmHsf01 in thermotolerance. PeerJ 8:e8926
Zhang X, Xu W, Ni D, Wang M, Guo G (2020) Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol 20:1–17
Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J et al (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368(3):515–521
Li Z, Zhang L, Wang A, Xu X, Li J (2013) Ectopic overexpression of SlHsfA3, a heat stress transcription factor from tomato, confers increased thermotolerance and salt hypersensitivity in germination in transgenic Arabidopsis. PLoS ONE 8(1):e54880
Li XD, Wang XL, Cai YM, Wu JH, Mo BT, Yu ER (2017) Arabidopsis heat stress transcription factors A2 (HSFA2) and A3 (HSFA3) function in the same heat regulation pathway. Acta Physiol Plant 39(3):67
Friedrich T, Oberkofler V, Trindade I, Altmann S, Brzezinka K, Lämke J et al (2021) Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat Commun 12(1):1–15
Baniwal SK, Chan KY, Scharf KD, Nover L (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282(6):3605–3613
Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432(2):203–207
Zhu WJ, Li PP, Xue CM, Chen M, Wang Z, Yang Q (2021) Potato plants overexpressing SpHsfA4c exhibit enhanced tolerance to high-temperature stress. Russ J Plant Physiol. https://doi.org/10.1134/S1021443721060248
Andrási N, Rigó G, Zsigmond L, Pérez-Salamó I, Papdi C, Klement E et al (2019) The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J Exp Bot 70(18):4903–4918
Du X, Li W, Sheng L, Deng Y, Wang Y, Zhang W et al (2018) Over-expression of chrysanthemum CmDREB6 enhanced tolerance of chrysanthemum to heat stress. BMC Plant Biol 18(1):1–10
Wang C, Zhou Y, Yang X, Zhang B, Xu F, Wang Y et al (2022) The heat stress transcription factor LlHsfA4 enhanced basic thermotolerance through regulating ROS metabolism in lilies (Lilium longiflorum). Int J Mol Sci 23(1):572
Huang YC, Niu CY, Yang CR, Jinn TL (2016) The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol 172(2):1182–1199
Kumar RR, Goswami S, Singh K, Dubey K, Rai GK, Singh B et al (2018) Characterization of novel heat-responsive transcription factor (TaHSFA6e) gene involved in regulation of heat shock proteins (HSPs)—a key member of heat stress-tolerance network of wheat. J Biotechnol 279:1–12
Poonia AK, Mishra SK, Sirohi P, Chaudhary R, Kanwar M, Germain H, Chauhan H (2020) Overexpression of wheat transcription factor (TaHsfA6b) provides thermotolerance in barley. Planta 252(4):1–14
Bi H, Zhao Y, Li H, Liu W (2020) Wheat heat shock factor TaHsfA6f increases ABA levels and enhances tolerance to multiple abiotic stresses in transgenic plants. Int J Mol Sci 21(9):3121
Seni S, Kaur S, Malik P, Yadav IS, Sirohi P, Chauhan H et al (2021) Transcriptome based identification and validation of heat stress transcription factors in wheat progenitor species Aegilops speltoides. Sci Rep 11(1):1–15
Sugio A, Dreos R, Aparicio F, Maule AJ (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21(2):642–654
Mesihovic A (2018) Regulation of the cellular response to elevated temperatures by heat stress transcription factor HsfA7 in “Solanum lycopersicum” (Doctoral dissertation, Johann Wolfgang Goethe-Universität Frankfurt am Main)
Mesihovic A, Ullrich S, Rosenkranz RR, Gebhardt P, Bublak D, Eich H, Fragkostefanakis S (2022) HsfA7 coordinates the transition from mild to strong heat stress response by controlling the activity of the master regulator HsfA1a in tomato. Cell reports 38(2):110224
Patel K, Bidalia A, Tripathi I, Gupta Y, Arora P, Rao KS (2021) Effect of heat stress on wild-type and A7a knockout mutant Arabidopsis thaliana plants. Vegetos. https://doi.org/10.1007/s42535-021-00272-4
Ikeda M, Mitsuda N, Ohme-Takagi M (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157(3):1243–1254
Fragkostefanakis S, Simm S, El-Shershaby A, Hu Y, Bublak D, Mesihovic A et al (2019) The repressor and co-activator HsfB1 regulates the major heat stress transcription factors in tomato. Plant Cell Environ 42(3):874–890
Rao S, Balyan S, Das JR, Verma R, Mathur S (2021) Transcriptional regulation of HSFA7 and post-transcriptional modulation of HSFB4a by miRNA4200 govern general and varietal thermotolerance in tomato. BioRxiv 7:1
Ma H, Wang C, Yang B, Cheng H, Wang Z, Mijiti A, Ma L (2016) CarHSFB2, a class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.). Plant Mol Biol Reporter 34(1):1–14
Hu XJ, Chen D, Lynne Mclntyre C, Fernanda Dreccer M, Zhang ZB, Drenth J et al (2018) Heat shock factor C2a serves as a proactive mechanism for heat protection in developing grains in wheat via an ABA-mediated regulatory pathway. Plant Cell Environ 41(1):79–98
Jiao SZ, Guo C, Yao WK, Zhang NB, Zhang JY, Xu WR (2022) An Amur grape VaHsfC1 is involved in multiple abiotic stresses. Sci Hortic 295:110785
Acknowledgements
The author wants to thank several colleagues for the scientific discussion, which helped improve the manuscript’s content. The author apologizes to all colleagues whose relevant work could not be cited due to space limitations.
Funding
This work receives no external funding.
Author information
Authors and Affiliations
Contributions
SH and TM conceived the idea. SH, MS, and AR wrote the manuscript. AR, SH, JI, and TM revised and improved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Haider, S., Raza, A., Iqbal, J. et al. Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal. Mol Biol Rep 49, 5771–5785 (2022). https://doi.org/10.1007/s11033-022-07190-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07190-x