Abstract
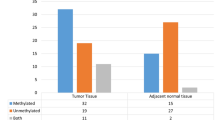
Promoter methylation mediated silencing of tumor suppressor genes plays an important role in the tumorigenesis of colorectal carcinoma (CRC). Tumor suppressor gene, Insulin-like Growth Factor Binding Protein-3 (IGFBP-3) expression is frequently downregulated in CRC due to promoter methylations. The aim of this study was to analyze the methylation status of IGFBP-3 gene promoter in stage II and III of CRC cases; find its association with clinicopathological characteristics of CRC patients and the methylation patterns as a prognostic biomarker. 58 histopathologically confirmed cases of CRC were included in the study. Methylation status of IGFBP-3 gene promoter was determined by using methylation specific PCR (MS-PCR) and bisulfite sequencing. Kaplan–Meier survival curve and univariate cox regression analysis were used for survival analysis; Chi-square test used for association analysis. IGFBP3 promoter methylation was found in 37 (63.8%) out of 58 CRC cases. This promoter methylation status was significantly associated with lymph-node metastasis (P = 0.013) and the survival period. In stage II CRC cases, unmethylated gene promoter status showed better survival than the methylated. Mean overall survival (OS) of methylated and unmethylated group was 22.23 months, and 49.15 months respectively (P = 0.045), HR = 6.432, 95% CI 0.986–41.943. The IGFBP-3 promoter methylations found in 63.8% CRC cases in this study. The methylations was found to be associated with lymph-node metastasis and overall survival of the patients particularly in stage II CRC patients. However, promoter methylation was not associated with other clinocopathological characteristics such as age, gender, tumor location etc.



Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Marzouk O, Schofield J (2011) Review of histopathological and molecular prognostic features in colorectal cancer. Cancers 3(2):2767–2810
Prabhu JS, Korlimarla A, Banerjee A, Wani S, Payal K, Sahoo R (2009) Gene-specific methylation: potential markers for colorectal cancer. Int J Biol Mark 24(1):57–62
Okugawa Y, Grady WM, Goel A (2015) Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149(5):1204–1225
Chand M, Keller DS, Mirnezami R, Bullock M, Bhangu A, Moran B et al (2018) Novel biomarkers for patient stratification in colorectal cancer: a review of definitions, emerging concepts, and data. World J Gastrointest Oncol 10(7):145
Weller M, Stupp R, Reifenberger G, Brandes AA, Van Den Bent MJ, Wick W, Hegi ME (2010) MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 6(1):39
Espada J, Esteller M (2007) Epigenetic control of nuclear architecture. Cell Mol Life Sci 64(4):449
Luczak MW, Jagodziński PP (2006) The role of DNA methylation in cancer development. Folia Histochem Cytobiol 44(3):143–154
Laird PW (2005) Cancer epigenetics. Hum Mol Genet 14:R65–R76
Revill K, Tim W, Anja L, Kensuke K, Andrew H, Jinyu L, Yujin H, Josep ML, Scott P (2013) Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 145(6):1424–1435
Tomii K, Tsukuda K, Toyooka S, Dote H, Hanafusa T, Asano H, Naitou M (2007) Aberrant promoter methylation of insulin-like growth factor binding protein-3 gene in human cancers. Int J Cancer 120(3):566–573
Albiston AL, Herington AC (1992) Tissue distribution and regulation of insulin-like growth factor (IGF)-binding protein-3 messenger ribonucleic acid (mRNA) in the rat: comparison with IGF-I mRNA expression. Endocrinology 130(1):497–502
Ricort JM (2004) Insulin-like growth factor binding protein (IGFBP) signalling. Growth Horm IGF Res 14(4):277
Hong J, Zhang G, Dong F, Rechler MM (2002) Insulin-like growth factor (IGF)-binding protein-3 mutants that do not bind IGF-I or IGF-II stimulate apoptosis in human prostate cancer cells. J Biol Chem 277(12):10489–10497
Kansra S, Ewton DZ, Wang J, Friedman E (2000) Igfbp-3 mediates TGFβ1 proliferative response in colon cancer cells. Int J Cancer 87(3):373–378
Perez-Carbonell L, Balaguer F, Toiyama Y, Egoavil C, Rojas E, Guarinos C, Andreu M et al (2014) IGFBP3 methylation is a novel diagnostic and predictive biomarker in colorectal cancer. PLoS ONE 9(8):e104285
Perks CM, Holly JM (2015) Epigenetic regulation of insulin-like growth factor binding protein-3 (IGFBP-3) in cancer. J Cell Commun Signal 9(2):159–166
Cortes-Sempere M, De Miguel MP, Pernia O, Rodriguez C, de Castro Carpeno J, Nistal MC et al (2013) IGFBP-3 methylation-derived deficiency mediates the resistance to cisplatin through the activation of the IGFIR/Akt pathway in non-small cell lung cancer. Oncogene 32(10):1274
Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S (2007) IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia (New York, NY) 9(12):1091
Torng P-L, Lin C-W, Chan MWY, Yang H-W, Huang S-C, Lin C-T (2009) Promoter methylation of IGFBP-3 and p53 expression in ovarian endometrioid carcinoma. Mol Cancer 8(1):120
Yi JM, Dhir M, Van Neste L, Downing SR, Jeschke J, Glockner SC, de Freitas CM, Hooker CM, Funes JM, Boshoff C et al (2011) Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res 17:1535–1545
Kim ST, Jang HL, Lee J, Park SH, Park YS, Lim HK et al (2015) Clinical significance of IGFBP-3 methylation in patients with early stage gastric cancer. Transl Oncol 8(4):288–294
Fu T, Pappou EP, Guzzetta AA, de Freitas Calmon M, Sun L, Herrera T et al (2016) IGFBP-3 gene methylation in primary tumor predicts recurrence of stage II colorectal cancers. Ann Surg 263(2):337
Herman JG, Graff JR, Myöhänen SBDN, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93(18):9821–9826
Susan JC, Harrison J, Paul CL, Frommer M (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22(15):2990–2997
Jogie-Brahim S, Feldman D, Oh Y (2009) Unraveling insulin-like growth factor binding protein-3 actions in human disease. Endocr Rev 30(5):417–437
Baxter RC (2014) IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer 14(5):329–341. https://doi.org/10.1038/nrc3720
Hanafusa T, Yumoto Y, Nouso K, Nakatsukasa H, Onishi T, Fujikawa T et al (2002) Reduced expression of insulin-like growth factor binding protein-3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett 176(2):149–158
Chang YS, Wang L, Liu D, Mao L, Hong WK, Khuri FR, Lee HY (2002) Correlation between insulin-like growth factor-binding protein-3 promoter methylation and prognosis of patients with stage I non-small cell lung cancer. Clin Cancer Res 8(12):3669–3675
Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, Monk D, Hata K, Marques-Bonet T, Wang L, Esteller M (2013) DNA methylation contributes to natural human variation. Genome Res 23:1363–1372
Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, Myers J, Godley LA, Dolan ME, Zhang W (2013) Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics 194:987–996
Fraser HB, Lam LL, Neumann SM, Kobor MS (2012) Population-specificity of human DNA methylation. Genome Biol 13:R8
Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM, MacIsaac JL et al (2018) Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol 19(1):1–17
Fraser HB, Lam LL, Neumann SM, Kobor MS (2012) Population-specificity of human DNA methylation. Genome Biol 13(2):1–12
Dar AA (2010) Functional modulation of insulin-like growth factor binding protein-3 in melanoma. Cell Mol Biol. https://doi.org/10.1158/1538-7445.AM10-5004
Georges RB, Adwan H, Hamdi H, Hielscher T, Linnemann U, Berger MR (2011) The insulin-like growth factor binding proteins 3 and 7 are associated with colorectal cancer and liver metastasis. Cancer Biol Ther 12(1):69–79
Gailhouste L, Liew LC, Hatada I, Nakagama H, Ochiya T (2018) Epigenetic reprogramming using 5-azacytidine promotes an anti-cancer response in pancreatic adenocarcinoma cells. Cell Death Dis 9(5):1–12
Dahn ML, Cruickshank BM, Jackson AJ, Dean C, Holloway RW, Hall SR, Coyle KM et al (2020) Decitabine response in breast cancer requires efficient drug processing and is not limited by multidrug resistance. Mol Cancer Ther 19(5):1110–1122
Acknowledgements
The authors would like to thank the funding agencies, University Grant commission (UGC), New Delhi, India, for UGC, National fellowship for SC (RGNF)—(F1-17.1-2017-18/RGNF-2017-18-SC-UTT-30798) and UGC-Non-Net fellowship-2013-17 for Mr. Alok Kumar. The authors also wish to thank Mr. Manoj Pandey for his help in the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declares that they have no conflict of interest.
Ethical approval
The study was ethically approved by the Institutional Ethics Committee (IEC), Dr. Ram Manohar Lohia Institute of Medical Science, Lucknow, Uttar Pradesh, India (IEC No.: IEC-8/15).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, A., Singh, P., Pandey, A. et al. IGFBP3 gene promoter methylation analysis and its association with clinicopathological characteristics of colorectal carcinoma. Mol Biol Rep 47, 6919–6927 (2020). https://doi.org/10.1007/s11033-020-05747-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05747-2