Abstract
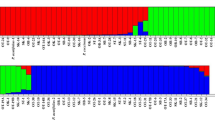
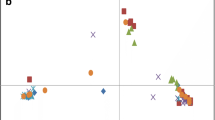
Common bean (Phaseolus vulgaris L.) is an important source of proteins, fibers and minerals for humans, being grown mainly in developing countries and representing a source of income for small farmers. In this work, a set of 206 Brazilian landraces and 59 elite lineages and cultivars were genotyped with 23 SSR (Simple Sequence Repeats) and 251 SNPs (Single-Nucleotide Polymorphism) markers. The ideal number of groups, according to STRUCTURE, was K = 2 for both SNPs and SSRs. This could be expected considering the two original gene pools—Andean (AND) and Mesoamerican (MES). The matrices of genetic simple matching dissimilarity for SSRs and SNPs were highly correlated; therefore, the allelic data of the markers was combined and analyzed to understand the genetic relationships of the studied collection. The neighbor-joining analysis considering the genetic distance of simple matching grouped the 265 genotypes into 17 subgroups. The markers SSR and SNP presented high power to discriminate among the genotypes. The ample genetic diversity observed in the work collection makes it a valuable source for the conservation, sustainable management and exploration in breeding programs of the crop.





Similar content being viewed by others
References
Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, Vanderleyden J (2003) Beans (Phaseolus spp.) – model food legumes. Plant Soil 252:55–128
Angioi LSA, Rau D, Attene G, Nanni L, Bellucci E, Logozzo G, Negri V, Spagnoletti Zeuli PL, Papa R (2010) Beans in Europe: origin and structure of the European landraces of Phaseolus vulgaris. Theor Appl Genet 121:829–843. https://doi.org/10.1007/s00122-010-1353-2
Castro-Rosas J, Aguirre-Santos EA, Gómez-Aldapa CA, Valle-Cervantes S, Ochoa-Martínez LA, Hernández-Santos B, Rodríguez-Miranda J (2016) Effect of harvest year on the physical properties, chemical composition and cooking time of three common bean varieties that are grown in Mexico. Qual Assur Saf Crops Foods 8:339–348
Santiago-Ramos D, Figueroa-Cárdenas JD, Véles-Medina JJ, Salazar R (2018) Physicochemical properties of nixtamalized black bean (Phaseolus vulgaris L.) flours. Food Chem 240:456–462
FAO. Faostat. Crops. (2017), https://www.fao.org/faostat/en/#data/QC (February 20, 2019)
Ramalho MAP, Abreu AFB (1998) Cultivares. In: Vieira C, Paula Junior TJ, Borém A (eds.) Feijão: aspectos gerais e cultura no Estado de Minas. Viçosa:UFV, pp 435–449
Schmutz J, McClean BE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713
Bitocchi E, Nanni L, Bellucci E, Rossi M, Giardini A, Zeuli PS, Logozzo G, Stougaard J, McClean P, Attene G et al (2012) Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc Natl Acad Sci USA 109:788–796
Gepts P (1988) A Middle American and an Andean common bean pool. In: Gepts P (ed) Genetic resources of Phaseolus beans. Kluwer Academic Publishers, Dordrecht, pp 375–390
Gepts P, Osborn TC, Rashka K, Bliss FA (1986) Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris): evidence for multiple centers of domestication. Econ Bot 40:451–468
Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor Appl Genet 118:979–992
Emygdio BM, Antunes IF, Nedel JL, Choer E (2003) Diversidade genética em cultivares locais e comerciais de feijão baseada em marcadores RAPD. Pesquisa Agropecuária Brasileira 38:1165–1171
Maciel FL, Echeverrigaray S, Gerald LTS, Grazziotin FG (2003) Genetic relationships and diversity among Brazilian cultivars and landraces of common beans (Phaseolus vulgaris L.) revealed by AFLP markers. Genet Resour Crop Evol 50:887–893
Perseguini JMKC, Oblessuc PR, Rosa JRBF, Gomes KA, Chiorato AF, Carbonell SAM, Garcia AAF, Vianello RP, Benchimol-Reis LL (2016) Genome-Wide Association Studies of Anthracnose and Angular Leaf Spot Resistance in Common Bean (Phaseolus vulgaris L.). PLoS ONE 11:1–19
Incaper (2016) Ações do Estado para a agricultura familiar é tema de reunião na Sede do Incaper, https://incaper.es.gov.br/Not%C3%ADcia/acoes-do-estado-para-a-agricultura-familiar-e-tema-de-reuniao-na-sede-do-incaper (March 1, 2018)
Cortés AJ, Chavarro MC, Blair MW (2011) SNP marker diversity in common bean (Phaseolus vulgaris L.). Theor Appl Genet 123:827–845
Müller BSF, Pappas GJ, Valdisser PAMR, Coelho GRC, Menezes IPP, Abreu AG, Borba TCO, Sakamoto T, Brondani C, Barros EG et al (2015) An operational SNP panel integrated to SSR marker for the assessment of genetic diversity and population structure of the common bean. Plant Mol Biol Rep 33:1697–1711
Mammadov J, Aggarwal R, Buyyarapu R, Kumpatla S (2012) SNP markers and their impact on plant breeding. Int J Plant Genom 2012:1–11
Song Q, Jia G, Hyten DL, Jenkins J, Hwang E-Y, Schroeder SG, Schmutz J, Jackson SA, McClean PE et al (2015) SNP assay development for linkage map construction, anchoring whole genome sequence and other genetic and genomic applications in common bean. G3 11:2285–2290
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Valdisser PAMR, Mota APS, Bueno LG, Menezes IPP, Coelho GRC, Magalhães FOC and Vianello (2013) Protocolo de extração de DNA e genotipagem de SSRs em larga escala para uso no melhoramento do feijoeiro comum (Phaseolus vulgaris L.). Publisher Embrapa, Santo Antônio de Goiás, 6 pp. (Technical Announcement, 208)
Cruz CD (2013) GENES - a software package for analysis in experimental statistics and quantitative genetics. Acta Sci 35(3):271–276
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Erence of population structure using multilocus genotype data. Genetics 155:945–959
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Earl DA, Vonholdta BM (2012) Website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Perrier X, Jacquemoud-Collet JP (2006) Darwin software, https://darwin.cirad.fr/darwin (March 1 2018)
Excoffier L, Heckel G (2006) Computer programs for population genetics data analysis: a survival guide. Nature 7:745–758
Wright S (1951) The genetical structure of populations. Ann Eugenics 15:223–354
Weir BS, Cockerham CC (1984) Estimating F-Statistics for the analysis of population structure. Evolution 38:1358–1370
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices. Version 2.9.3, https://www2.unil.ch/popgen/softwares/fstat.htm (March 1, 2018)
Burle ML, Fonseca JR, Kami JA, Gepts P (2010) Microsatellite diversity and genetic structure among common bean (Phaseolus vulgaris L.) landraces in Brazil, a secondary center of diversity. Theor Appl Genet 121:801–813
Valdisser PAMR, Pappas GJ Jr, Menezes IPP, Müller BSF, Pereira WJ, Narciso MG, Brondani C, Souza TLPO, Borba TCO, Vianello RP (2016) SNP discovery in common bean by restriction-associated DNA (RAD) sequencing for genetic diversity and population structure analysis. Mol Genet Genomics 291:1277–1291
Valdisser PAMR, Pereira WJ, Almeida Filho JE, Müller BSF, Coelho GRC, Menezes IPP, Vianna JPG, Zucchi MI, Lanna AC, Coelho ASG et al (2017) In-depth genome characterization of a Brazilian common bean core collection using DArTseq high-density SNP genotyping. BMC Genomics 18:423
Blair MW, Brondani RVP, Díaz LM, Peloso MJ (2013) Diversity and population structure of common bean from Brazil. Crop Sci 53:1983–1993
Cardoso PCB, Brondani C, Menezes IPP, Valdisser PAMR, Borba TCO, Del Peloso MJ, Vianello RP (2014) Discrimination of common bean cultivars using multiplexed microsatellite markers. Genet Mol Res 13:1964–1978
Del Peloso MJ and Melo LC (2005) Potencial de rendimento da cultura do feijoeiro comum. Santo Antônio de Goiás: Embrapa Arroz e Feijão
Benin G, Carvalho FIF, Assmann IC, Cigolini J, Cruz PJ, Machioro VS, Lorencetti C, Silva JAG (1955) Identificação da dissimilaridade genética entre genótipos de feijoeiro comum (Phaseolus vulgaris L.) do grupo preto. Curr Agric Sci Technol 8:179–184
Pereira HS, Melo LC, Del Peloso MJ, Díaz JLC, Magaldi MCS, Faria LC, Abreu AFB, Pereira Filho IA, Moreira JAA, Martins M, Wendland A, Costa JGC (2010) Evaluation of export common bean genotypes in Brazil. Ann Rep Bean Improv Coop 53:276–277
Ceccarelli S (2014) Drought. In: Jackson M, Ford-Lloyd B, Parry M (eds) Plantgenetic resources and climate change. CAB International, Wallingford, pp 221–235
Blair MW, Iriarte G, Beebe S (2006) QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean× wild common bean (Phaseolus vulgaris L.) cross. Theor Appl Genet 112:1149–1163
Bitocchi E, Bellucci E, Giardini A, Rau D, Rodriguez M, Biagetti E, Santilocchi R, Zeuli OS, Gioia T, Logozzo G et al (2013) Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol 197:300–313
Mamidi S, Rossi M, Annam D, Moghaddam S, Lee R, Papa R, McClean PE (2011) Investigation of the domestication of common bean (Phaseolus vulgaris) using multilocus sequence data. Funct Plant Biol 38:953–967
Mamidi S, Rossi M, Moghaddam SM, Annam D, Lee R, Papa R, McClean PE (2013) Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 110:267–276
Acknowledgments
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), Fundação de Amparo à Pesquisa do Espírito Santo (FAPES, Vitória, ES, Brazil), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil) – Finance Code 001, for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5726_MOESM1_ESM.tif
Figure S1. Distribution of the polymorphism information content (PIC) values in the 217 SNP(left) loci and 23 SSR (right) loci used in the analyses of genetic diversity in the 249 common bean genotypes. (TIF 1320 kb)
11033_2020_5726_MOESM2_ESM.tif
Figure S2. Relationship between the sets of SNP (axis y) and SSR markers (axis x) calculated using simple matching dissimilarity (Mantel r = 0.7418). Each point represents one comparison between dissimilarity matrices. (TIF 2286 kb)
11033_2020_5726_MOESM3_ESM.tiff
Figure S3. Representative images demonstrating the diversity in size, shape and color of the bean collection analyzed in this study. (TIFF 2367 kb)
11033_2020_5726_MOESM7_ESM.docx
Table S4 – The 265 bean accessions clustered in the 17 groups generated by neighbor-joining analysis with the combination of 23 SSRs and 251 SNPs. (DOCX 31 kb)
Rights and permissions
About this article
Cite this article
Carvalho, M.S., de Oliveira Moulin Carias, C.M., Silva, M.A. et al. Genetic diversity and structure of landrace accessions, elite lineages and cultivars of common bean estimated with SSR and SNP markers. Mol Biol Rep 47, 6705–6715 (2020). https://doi.org/10.1007/s11033-020-05726-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05726-7