Abstract
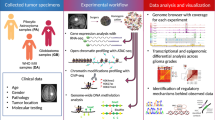
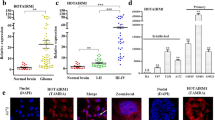
The long noncoding RNA HOTAIRM1 reportedly plays important roles in acute myeloid leukemia, gastric cancer and colorectal cancer. Here, we analyzed potential function of HOTAIRM1 in glioma and asked whether it participates in long-range chromatin interactions. We monitored expression of HOTAIRM1 in glioma tissues and correlated levels with patient survival using the TCGA dataset. HOTAIRM1 was highly expressed in glioma tissue, with high levels associated with shortened patient survival time. We then suppressed HOTAIRM1 activity in the human glioblastoma U251 line using CRISPR-cas9 to knock in a truncating polyA fragment. Reporter analysis of these and control cells confirmed that the HOTAIRM1 locus serves as an active enhancer. We then performed Capture-C analysis to identify target genes of that locus and applied RNA antisense purification to assess chromatin interactions between the HOTAIRM1 locus and HOXA cluster genes. HOTAIRM1 knockdown in glioma cells decreased proliferation and reduced expression of HOXA cluster genes. HOTAIRM1 regulates long-range interactions between the HOTAIRM1 locus and HOXA genes. Our work suggests a new mechanism by which HOTAIRM1 regulates glioma progression by regulating high-order chromatin structure and could suggest novel therapeutic targets to treat an intractable cancer.





Similar content being viewed by others
References
Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA (2001) Malignant glioma: genetics and biology of a grave matter. Genes Dev 15(11):1311–1333
Beiko J, Suki D, Hess KR, Fox BD, Cheung V, Cabral M, Shonka N, Gilbert MR, Sawaya R et al (2014) IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol 16(1):81–91
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172(3):393–407
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927
Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482:339–346
Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J et al (2005) Antisense transcription in the mammalian transcriptome. Science 309:1564–1566
Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46
Chen LL (2016) Linking long noncoding RNA localization and function. Trends Biochem Sci 41(9):761–772
Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I et al (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142(3):409–419
Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106(28):11667–11672
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147:358–369
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P et al (2011) Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 147(2):344–357
Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG et al (2017) mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541:228–232
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X, Sun T, Xie X, Zhou Y et al (2016) Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis 7:e2123
Xu LM, Chen L, Li F, Zhang R, Li ZY, Chen FF, Jiang XD (2016) Over-expression of the long non-coding RNA HOTTIP inhibits glioma cell growth by BRE. J Exp Clin Cancer Res 35(1):162
Wang G, Li Z, Tian N, Han L, Fu Y, Guo Z, Tian Y (2016) miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol Lett 12(2):879–886
Liu C, Zhang Y, She X, Fan L, Li P, Feng J, Fu H, Liu Q, Liu Q et al (2018) A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol 11(1):77–77
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539(7629):452–455
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W et al (2016) Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18:637–652
Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ (2017) The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ 24:212–224
Wei S, Zhao M, Wang X, Li Y, Wang K (2016) PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol 9:44
Diaz-Beya M, Brunet S, Nomdedeu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I et al (2015) The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 6:31613–31627
Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H (2016) HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med 20:2036–2044
Wang XQD, Dostie J (2017) Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res 45(3):1091–1104
Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292
Bickmore WA (2013) The spatial organization of the human genome. Ann Rev Genomics Hum Genet 14:67–84
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376
Rao Suhas SP, Huntley Miriam H, Durand Neva C, Stamenova Elena K, Bochkov Ivan D, Robinson James T, Sanborn Adrian L, Machol I, Omer Arina D et al (2015) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 162(3):687–688
Chen X, Zhang M, Gan H, Wang H, Lee J-H, Fang D, Kitange GJ, He L, Hu Z et al (2018) A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun 9(1):2949
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE (2015) Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529:110
Roy SS, Mukherjee AK, Chowdhury S (2018) Insights about genome function from spatial organization of the genome. Hum Genomics 12(1):8
Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494:497
Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, Lu C, Jia W, Xi J et al (2018) A linc1405/eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell 22(6):893–908
Davies JO, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, Hughes JR (2016) Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods 13(1):74–80
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A et al (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341(6147):1237973
Li Q, Dong C, Cui J, Wang Y, Hong X (2018) Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res 37(1):265
Song L, Zhang S, Duan C, Ma S, Hussain S, Wei L, Chu M (2019) Genome-wide identification of lncRNAs as novel prognosis biomarkers of glioma. J Cell Biochem 120(12):19518–19528
Liang Q, Li X, Guan G, Xu X, Chen C, Cheng P, Cheng W, Wu A (2019) Long non-coding RNA, HOTAIRM1, promotes glioma malignancy by forming a ceRNA network. Aging (Albany NY) 11(17):6805–6838
Pradeepa MM, McKenna F, Taylor GCA, Bengani H, Grimes GR, Wood AJ, Bhatia S, Bickmore WA (2017) Psip1/p52 regulates posterior Hoxa genes through activation of lncRNA Hottip. PLoS Genet 13(4):e1006677–e1006677
Orom UA, Shiekhattar R (2013) Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154(6):1190–1193
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J et al (2014) Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 24(5):513–531
Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I (2015) LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell 60(4):626–636
Cimino PJ, Kim Y, Wu HJ, Alexander J, Wirsching HG, Szulzewsky F, Pitter K, Ozawa T, Wang J et al (2018) Increased HOXA5 expression provides a selective advantage for gain of whole chromosome 7 in IDH wild-type glioblastoma. Genes Dev 32(7–8):512–523
Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, Zardo G, Nigro J, James CD et al (2010) Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res 70(2):453–462
Zang L, Kondengaden SM, Che F, Wang L, Heng X (2018) Potential epigenetic-based therapeutic targets for glioma. Front Mol Neurosci 11:408–408
Romani M, Pistillo MP, Banelli B (2018) Epigenetic Targeting of Glioblastoma. Front Oncol 8:448
Taylor LP (2010) Diagnosis, treatment, and prognosis of glioma: five new things. Neurology 75:S28–S32
Tian X, Ma J, Wang T, Tian J, Zhang Y, Mao L, Xu H, Wang S (2018) Long non-coding RNA HOXA transcript antisense RNA myeloid-specific 1-HOXA1 axis downregulates the immunosuppressive activity of myeloid-derived suppressor cells in lung cancer. Front Immunol 9:473
Quinonez SC, Innis JW (2014) Human HOX gene disorders. Mol Genet Metab 111(1):4–15
Bhatlekar S, Fields JZ, Boman BM (2014) HOX genes and their role in the development of human cancers. J Mol Med 92(8):811–823
Collins CT, Hess JL (2016) Role of HOXA9 in leukemia: dysregulation, cofactors and essential targets. Oncogene 35(9):1090–1098
Taminiau A, Draime A, Tys J, Lambert B, Vandeputte J, Nguyen N, Renard P, Geerts D, Rezsohazy R (2016) HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-kappaB pathway in a transcription-independent manner. Nucleic Acids Res 44(15):7331–7349
Cheng S, Qian F, Huang Q, Wei L, Fu Y, Du Y (2018) HOXA4, down-regulated in lung cancer, inhibits the growth, motility and invasion of lung cancer cells. Cell Death Dis 9(5):465
Di Vinci A, Casciano I, Marasco E, Banelli B, Ravetti GL, Borzi L, Brigati C, Forlani A, Dorcaratto A et al (2012) Quantitative methylation analysis of HOXA3, 7, 9, and 10 genes in glioma: association with tumor WHO grade and clinical outcome. J Cancer Res Clin Oncol 138(1):35–47
Shen X, Bai H, Zhu H, Yan Q, Yang Y, Yu W, Shi Q, Wang J, Li J et al (2018) Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate HOXA11 expression by sponging miR-181a in multiple myeloma. Cell Physiol Biochem 49(1):87–100
Chang S, Liu J, Guo S, He S, Qiu G, Lu J, Wang J, Fan L, Zhao W et al (2016) HOTTIP and HOXA13 are oncogenes associated with gastric cancer progression. Oncol Rep 35(6):3577–3585
Furlong EEM, Levine M (2018) Developmental enhancers and chromosome topology. Science 361(6409):1341
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31701129, 81772687, 31530027) and the Natural Science Foundation of Tianjin City of China (Grant No. 18JCQNJC10100).
Author information
Authors and Affiliations
Contributions
WL, LZ, and TS conceived the projects. TS, HX, GS, JC, ZZ, and JS performed the experiments. DG contributed to sequencing and bioinformatics analysis. TS, LZ, FA, and MW wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2020_5371_MOESM1_ESM.pdf
Supplementary Fig. 1— HOTAIRM1 downregulation inhibits glioma proliferation. A Kaplan-Meir survival curve of patients with high versus low HOTAIRM1 expression. Bars indicate s.e.m., ***p < 0.001. B Schematic of CRISPR-Cas9-mediated HOTAIRM1 polyA knock-in. C Efficiency of HOTAIRM1 knock-down in two glioma lines (KI-1 and KI-2). D Proliferation of indicated glioma lines, as determined by an MTS assay. E, G Colony formation assays of indicated lines. F,H Flow cytometry analysis of the cell cycle indicates that HOTAIRM1 knockdown is associated with a decreased number of cells in S and an increase in the number of cells in G1 or G2. Bars indicate s.e.m., ***p<0.001 (PDF 294 kb)
Rights and permissions
About this article
Cite this article
Shi, T., Guo, D., Xu, H. et al. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol Biol Rep 47, 2723–2733 (2020). https://doi.org/10.1007/s11033-020-05371-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05371-0