Abstract
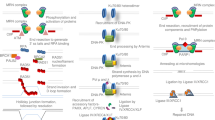
The majority of CRISPR-Cas9 methods for mutations correction are oriented on gene editing through homologous recombination that is normally restrained by non-homologous end joining (NHEJ). A recently identified protein TIRR can bind a 53BP1 protein, a key effector of NHEJ, and inhibit its recruitment to double-strand break loci. Several studies elucidated the molecular mechanisms of TIRR-53BP1 binding and established bidirectional role of TIRR in 53BP1 functions and stability. It was proved that overexpression of TIRR promotes the double-strand break repair through homologous recombination. All findings, which were described in the review, allow assuming TIRR as a suitable target for enhancing efficacy of genome editing through homology directed repair.




Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Change history
04 January 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11033-020-06098-8
References
Liu M, Rehman S, Tang X et al (2018) Methodologies for Improving HDR Efficiency. Front Genet 9:691. https://doi.org/10.3389/fgene.2018.00691
Wang M, Wu W, Wu W et al (2006) PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 34:6170–6182. https://doi.org/10.1093/nar/gkl840
Pawelczak KS, Gavande NS, VanderVere-Carozza PS, Turchi JJ (2018) Modulating DNA repair pathways to improve precision genome engineering. ACS Chem Biol 13:389–396. https://doi.org/10.1021/acschembio.7b00777
Pannunzio NR, Watanabe G, Lieber MR (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem 293:10512–10523. https://doi.org/10.1074/jbc.TM117.000374
Gutschner T, Haemmerle M, Genovese G et al (2016) Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep 14:1555–1566. https://doi.org/10.1016/j.celrep.2016.01.019
Chu VT, Weber T, Wefers B et al (2015) Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol 33:543–548. https://doi.org/10.1038/nbt.3198
Song J, Yang D, Xu J et al (2016) RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat Commun 7:10548. https://doi.org/10.1038/ncomms10548
Charpentier M, Khedher AHY, Menoret S et al (2018) CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat Commun 9:1133. https://doi.org/10.1038/s41467-018-03475-7
Byrne SM, Ortiz L, Mali P et al (2015) Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res 43:e21–e21. https://doi.org/10.1093/nar/gku1246
Zhang J-P, Li X-L, Li G-H et al (2017) Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol 18:35. https://doi.org/10.1186/s13059-017-1164-8
Richardson CD, Ray GJ, DeWitt MA et al (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34:339–344. https://doi.org/10.1038/nbt.3481
Ma D, Xu Z, Zhang Z et al (2019) Engineer chimeric Cas9 to expand PAM recognition based on evolutionary information. Nat Commun 10:560. https://doi.org/10.1038/s41467-019-08395-8
Smirnikhina SA, Anuchina AA, Lavrov AV (2019) Ways of improving precise knock-in by genome-editing technologies. Hum Genet 138:1–19. https://doi.org/10.1007/s00439-018-1953-5
Drané P, Brault M-E, Cui G et al (2017) TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 543:211–216. https://doi.org/10.1038/nature21358
Botuyan MV, Lee J, Ward IM et al (2006) Structural basis for the methylation state-specific recognition of histone H4–K20 by 53BP1 and Crb2 in DNA repair. Cell 127:1361–1373. https://doi.org/10.1016/j.cell.2006.10.043
Zhang A, Peng B, Huang P et al (2017) The p53-binding protein 1-Tudor-interacting repair regulator complex participates in the DNA damage response. J Biol Chem 292:6461–6467. https://doi.org/10.1074/jbc.M117.777474
Drané P, Chowdhury D (2017) TIRR and 53BP1-partners in arms. Cell Cycle 16:1235–1236. https://doi.org/10.1080/15384101.2017.1337966
Panier S, Boulton SJ (2014) Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 15:7–18. https://doi.org/10.1038/nrm3719
Fradet-Turcotte A, Canny MD, Escribano-Díaz C et al (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499:50–54. https://doi.org/10.1038/nature12318
Callen E, Faryabi RB, Luckey M et al (2012) The DNA damage- and transcription-associated protein paxip1 controls thymocyte development and emigration. Immunity 37:971–985. https://doi.org/10.1016/j.immuni.2012.10.007
Chapman JR, Barral P, Vannier J-B et al (2013) RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49:858–871. https://doi.org/10.1016/j.molcel.2013.01.002
Noordermeer SM, Adam S, Setiaputra D et al (2018) The shieldin complex mediates 53BP1-dependent DNA repair. Nature 560:117–121. https://doi.org/10.1038/s41586-018-0340-7
Findlay S, Heath J, Luo VM et al (2018) SHLD2/FAM35A co-operates with REV7 to coordinate DNA double-strand break repair pathway choice. EMBO J. https://doi.org/10.15252/embj.2018100158
Ghezraoui H, Oliveira C, Becker JR et al (2018) 53BP1 cooperation with the REV7–shieldin complex underpins DNA structure-specific NHEJ. Nature 560:122–127. https://doi.org/10.1038/s41586-018-0362-1
Reid DA, Keegan S, Leo-Macias A et al (2015) Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc Natl Acad Sci U S A 112:E2575–E2584. https://doi.org/10.1073/pnas.1420115112
Clawson GA, Abraham T, Pan W et al (2017) A cholecystokinin B receptor-specific DNA aptamer for targeting pancreatic ductal adenocarcinoma. Nucleic Acid Ther 27:23–35. https://doi.org/10.1089/nat.2016.0621
Lieber MR (2008) The mechanism of human nonhomologous DNA End joining. J Biol Chem 283:1–5
Cottarel J, Frit P, Bombarde O et al (2013) A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 200:173–186. https://doi.org/10.1083/jcb.201203128
Isono M, Niimi A, Oike T et al (2017) BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep 18:520–532. https://doi.org/10.1016/j.celrep.2016.12.042
Schlegel BP, Jodelka FM, Nunez R (2006) BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res 66:5181–5189. https://doi.org/10.1158/0008-5472.CAN-05-3209
Kijas AW, Lim YC, Bolderson E et al (2015) ATM-dependent phosphorylation of MRE11 controls extent of resection during homology directed repair by signalling through Exonuclease 1. Nucleic Acids Res 43:8352–8367. https://doi.org/10.1093/nar/gkv754
Shibata A, Moiani D, Arvai AS et al (2014) DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53:7–18. https://doi.org/10.1016/j.molcel.2013.11.003
Garcia V, Phelps SEL, Gray S, Neale MJ (2011) Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479:241–244. https://doi.org/10.1038/nature10515
Nimonkar AV, Genschel J, Kinoshita E et al (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25:350–362. https://doi.org/10.1101/gad.2003811
Butler LR, Densham RM, Jia J et al (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J 31:3918–3934. https://doi.org/10.1038/emboj.2012.232
Nakada S (2016) Opposing roles of RNF8/RNF168 and deubiquitinating enzymes in ubiquitination-dependent DNA double-strand break response signaling and DNA-repair pathway choice. J Radiat Res 57(Suppl 1):i33–i40. https://doi.org/10.1093/jrr/rrw027
Kakarougkas A, Ismail A, Katsuki Y et al (2013) Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res 41:10298–10311. https://doi.org/10.1093/nar/gkt802
Pellegrini L, Yu DS, Lo T et al (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420:287–293. https://doi.org/10.1038/nature01230
Kakarougkas A, Jeggo PA (2014) DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. https://doi.org/10.1259/bjr.20130685
Verma P, Greenberg RA (2016) Noncanonical views of homology-directed DNA repair. Genes Dev 30:1138–1154. https://doi.org/10.1101/gad.280545.116
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. https://doi.org/10.1146/annurev-genet-110410-132435
Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC (2008) Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem 283:14883–14892. https://doi.org/10.1074/jbc.M801097200
Ceccaldi R, Rondinelli B, D’Andrea AD (2016) Repair pathway choices and consequences at the double-strand break. Trends Cell Biol 26:52–64
Dabin J, Fortuny A, Polo SE (2016) Epigenome maintenance in response to DNA damage. Mol Cell 62:712–727. https://doi.org/10.1016/j.molcel.2016.04.006
Russo G, Landi R, Pezone A et al (2016) DNA damage and Repair Modify DNA methylation and chromatin domain of the targeted locus: mechanism of allele methylation polymorphism. Sci Rep 6:33222. https://doi.org/10.1038/srep33222
Aymard F, Bugler B, Schmidt CK et al (2014) Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol 21:366–374. https://doi.org/10.1038/nsmb.2796
Fnu S, Williamson EA, De Haro LP et al (2011) Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA 108:540–545. https://doi.org/10.1073/pnas.1013571108
Wei S, Li C, Yin Z et al (2018) Histone methylation in DNA repair and clinical practice: new findings during the past 5-years. J Cancer 9:2072–2081. https://doi.org/10.7150/jca.23427
Simonetta M, de Krijger I, Serrat J et al (2018) H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle 17:124–136. https://doi.org/10.1080/15384101.2017.1404210
Baciu PC, Saoncella S, Lee SH et al (2000) Syndesmos, a protein that interacts with the cytoplasmic domain of syndecan-4, mediates cell spreading and actin cytoskeletal organization. J Cell Sci 113(Pt 2):315–324
Denhez F, Wilcox-Adelman SA, Baciu PC et al (2002) Syndesmos, a syndecan-4 cytoplasmic domain interactor, binds to the focal adhesion adaptor proteins paxillin and Hic-5. J Biol Chem 277:12270–12274. https://doi.org/10.1074/jbc.M110291200
Srouji JR, Xu A, Park A et al (2017) The evolution of function within the Nudix homology clan. Proteins 85:775–811. https://doi.org/10.1002/prot.25223
McLennan AG (2006) The Nudix hydrolase superfamily. Cell Mol Life Sci 63:123–143. https://doi.org/10.1007/s00018-005-5386-7
Bessman MJ, Frick DN, O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271:25059–25062. https://doi.org/10.1074/jbc.271.41.25059
National Center of Biotechnology Information (2019) NUDT16L1 nudix hydrolase 16 like 1 [Homo sapiens (human)]. https://www.ncbi.nlm.nih.gov/gene/84309. Accessed 05 Feb 2020
Iyama T, Abolhassani N, Tsuchimoto D et al (2010) NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res 38:4834–4843. https://doi.org/10.1093/nar/gkq249
Taylor MJ, Peculis BA (2008) Evolutionary conservation supports ancient origin for NUDT16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res 36:6021–6034. https://doi.org/10.1093/nar/gkn605
Dai Y, Zhang A, Shan S et al (2018) Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR. Nat Commun 9:2123. https://doi.org/10.1038/s41467-018-04557-2
Wang J, Yuan Z, Cui Y et al (2018) Molecular basis for the inhibition of the methyl-lysine binding function of 53BP1 by TIRR. Nat Commun 9:2689. https://doi.org/10.1038/s41467-018-05174-9
Schrödinger, Inc. (2018) PyMOL Molecular Graphics System. https://pymol.org/2/
Tangutoori S, Baldwin P, Sridhar S (2015) PARP inhibitors: A new era of targeted therapy. Maturitas 81:5–9. https://doi.org/10.1016/j.maturitas.2015.01.015
Javle M, Curtin NJ (2011) The role of PARP in DNA repair and its therapeutic exploitation. Br J Cancer 105:1114–1122. https://doi.org/10.1038/bjc.2011.382
Botuyan MV, Cui G, Drané P et al (2018) Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein. Nat Struct Mol Biol. https://doi.org/10.1038/s41594-018-0083-z
Avolio R, Järvelin AI, Mohammed S et al (2018) Protein syndesmos is a novel RNA-binding protein that regulates primary cilia formation. Nucleic Acids Res 46:12067–12086. https://doi.org/10.1093/nar/gky873
Hu Z, Shi Z, Guo X et al (2018) Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR–Cas9 and ssODN in human cancer cells. Cell Biosci 8:12. https://doi.org/10.1186/s13578-018-0200-z
Gerlach M, Kraft T, Brenner B et al (2018) Efficient Knock-in of a point mutation in porcine fibroblasts using the CRISPR/Cas9-GMNN Fusion gene. Genes (Basel) 9:296. https://doi.org/10.3390/genes9060296
Pinder J, Salsman J, Dellaire G (2015) Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res 43:9379–9392. https://doi.org/10.1093/nar/gkv993
Robert F, Barbeau M, Éthier S et al (2015) Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med 7:93. https://doi.org/10.1186/s13073-015-0215-6
Yang D, Scavuzzo MA, Chmielowiec J et al (2016) Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases. Sci Rep 6:21264. https://doi.org/10.1038/srep21264
Ye L, Wang C, Hong L et al (2018) Programmable DNA repair with CRISPRa/i enhanced homology-directed repair efficiency with a single Cas9. Cell Discov 4:46. https://doi.org/10.1038/s41421-018-0049-7
Canny MD, Moatti N, Wan LCK et al (2017) Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR–Cas9 genome-editing efficiency. Nat Biotechnol 36:95–102. https://doi.org/10.1038/nbt.4021
Acknowledgements
The section “The interplay of the main factors managing repair of DNA DSB” was supported by the grant of the Russian Science Foundation (Agreement 17-75-20095), the section “The role of chromatin condition in the repair pathway choice” was supported by the Russian Academy of Sciences (Program “Fundamental researches for biomedical technologies”). The results of parts “The first histone-masking protein” and “TIRR role in DNA repair” were obtained within the state assignment of Ministry of Science and Higher Education of Russian Federation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Anuchina, A.A., Lavrov, A.V. & Smirnikhina, S.A. TIRR: a potential front runner in HDR race−hypotheses and perspectives. Mol Biol Rep 47, 2371–2379 (2020). https://doi.org/10.1007/s11033-020-05285-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05285-x