Abstract
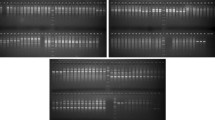
The genus of Bromus is one of the most important collection of rangeland plants, which are distributed in a wide range of natural areas of Iran. Interspecific relationships were evaluated in 90 accessions of 18 Bromus species based on 15 ISSR and 15 SCoT primers. SCoT markers separated the accessions better than ISSR marker. In addition, there was a high interspecific diversity between surveying germplasm. The sections of Bromus genus completely separated based on DNA molecular markers. SCoT markers could separate the accessions in each species. The primers of SC5 and SC35 from SCoT marker and UBC861, UBC857 and UBC844 primers from ISSR marker were identified as the best primers in revealing of genetic diversity between accessions. The sections of Ceratochloa, Genea, Pnigma and Bromus were monophyletic and were placed in one cluster. The section Bromus had a direct relationship with section Genea. In other words, section Ceratochloa has a direct relationship with Pnigma. B. tectorum and B. sericeus. B. sterilis had the most distance with other species in section Genea. B. squarrosus and B. japonicus had the most similarity and B. briziformis with B. danthoniae and B. scoparius with B. rechingeri had a moderate relationship in section Bromus. B. tomentosus and B. persicus had the highest similarity and B. riparius with B. biebersteinii and B. tomentellus with B. inermis had a moderate similarity in section Pnigma.








Similar content being viewed by others
References
Arzani A, Kaligi MR, Shiran B, Kharazian N (2005) Evaluation of diversity in wild relatives of wheat. Czech J Genet Plant Breading 41:112–117
Williams WM, Stewart AV, Williamson ML (2011) Bromus. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, millets and grasses. Springer, New York
Sanderson MA, Skinner RH, Elwinger GF (2002) Seedling development and field performance of prairiegrass, grazing brome-grass, and orchardgrass. Crop Sci 42:224–230
Gould FW, Shaw RB (1983) Grass systematics. Texas University Press, College Station
Soderstrom TR, Beaman JH (1968) The genus Bromus (Gramineae) in Mexico and Central America. Publ Mus Michigan State Univ Biol Ser 3:465–520
Saarela JM, Peterson P, Valdés-Reyna J (2014) A taxonomic revision of Bromus (Poaceae: Pooideae: Bromeae) in México and Central America. Phytotaxa 185(1):1–147
Naderi R, Rahiminejad MR (2015) A taxonomic revision of the genus Bromus (Poaceae) and a new key to the tribe Bromeae in Iran. Annales Botanici Fennici 52:233–248
Saarela JM, Peterson PM, Keane RM, Cayouette J, Graham SW (2007) Molecular phylogenetics of Bromus (Poaceae: Pooideae) based on chloroplast and nuclear DNA sequence data. J Syst Evol Bot 23(1):450–467
Aryavand A (2002) Phenetic analysis of the Iranian species of the Bromus sections Genea, Neobromus and Nevskiella. J Sci Islam Republ Iran 13(1):3–13
Bor NL (1970) Graminae. In: Rechinger KH (ed) Flora Iranica. Akademische Druk-U. Verlagsanstalt, Graz, pp 203–211
Sales F (1994) Evolutionary tendencies in some annual species of Bromus (Bromus L. sect. Genea Dum. (Poaceae)). Bot J Linn Soc 115:197–210
Moradkhani H, Mehrabi AA, Etminan A, Pour-Aboughadareh A (2015) Molecular diversity and phylogeny of Triticum-Aegilops species possessing D genome revealed by SSR and ISSR markers. Plant Breeding Seed Sci 71:81–95
Safari H, Shirvani H, Jafari AA, Mahdavi S (2014) The study of genetic variation for Lolium perenne using ISSR molecular markers. Int J Biosci 4(1):75–81
Rouf MA, Andrew AH, John CZ (2002) Determination of genetic diversity in Tall Fescue with AFLP markers. Crop Sci 42:944–950
Collard BCY, Mackill DJ (2009) Start Codon Targeted (SCoT) polymorphism: a simple novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol 27:86–93
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen J, Hyvonen J (2013) Advances in plant gene-targeted and functional markers. Plant Methods 9(6):31p
Yang S, Xue Sh, Kang W, Qian Z, Yi Z (2019) Genetic diversity and population structure of Miscanthus lutarioriparius, an endemic plant of China. PLoS ONE 14(2):e0211471
Ainouche ML, Bayer RJ (1997) On the origins of the tetraploid Bromus species (section Bromus, Poaceae): insights from the internal transcribed spacer sequences of nuclear ribosomal DNA. Genome 40:730–743
Ainouche ML, Bayer RJ, Gourret JP, Defontaine A, Misset MT (1999) The allotetraploid invasive weed Bromus hordeaceus L. (Poaceae): genetic diversity, origin and molecular evolution. Folia Geobot 34:405–419
Fortune PM, Pourtau N, Viron N, Ainouche ML (2008) Molecular phylogeny and reticulate origins of the polyploid Bromus species from section Genea (Poaceae). Am J Bot 95(4):454–464
Pillay M (1995) Chloroplast DNA similarity of smooth bromegrass with other pooid cereals: implications for plant breeding. Crop Sci 35:869–875
Pillay M, Hilu KW (1995) Chloroplast-DNA restriction site analysis in the genus Bromus (Poaceae). Am J Bot 82:239–249
Pillay M, Armstrong KC (2001) Maternal inheritance of chloroplast DNA in interspecific crosses of Bromus. Biol Plant 44:47–51
Joachimiak A, Sutkowska A, Mitka J (2001) RAPD studies in Bromus (Poaceae) from the old and new worlds—preliminary results. Acta Biol Acad Sci Hung. 43:79–86
Sutkowska A, Mitka J (2008) RAPD analysis points to old world Bromus species as ancestral to new world subgen, Festucaria. Acta Biol Crac Bot 50:117–125
Massa AN, Larson SR, Jensen KB, Hole DJ (2001) AFLP variation in Bromus section Ceratochloa germplasm of Patagonia. Crop Sci 41:1609–1616
Fu YB, Coulman BE, Ferdinandez YSN, Cayouette J, Peterson PM (2005) Genetic diversity of fringed brome (Bromus ciliatus) as determined by amplified fragment length polymorphism. Can J Bot 83:1322–1328
Sutkowska A, Pasierbiński A, Warzecha T, Mandal A, Mitka J (2013) Refugial pattern of Bromus erectus in central Europe based on issr fingerprinting. Acta Biol Crac Ser Bot 55(2):107–119
Sutkowska A, Pasierbiński A, Warzecha T, Mitka J (2014) Multiple cryptic refugia of forest grass Bromus benekenii in Europe as revealed by ISSR fingerprinting and species distribution modelling. Plant Syst Evol 300:1437–1452
Sutkowska A, Pasierbiński A, Bąba W (2015) Additivity of ISSR markers in natural hybrids of related forest species Bromus benekenii and B. ramosus (Poaceae). Acta Biol Crac Ser Bot 57(1):82–94
Sutkowska A (2012) Verification of the systematic position of California Brome (Bromus carinatus Hook. and Arn., Poaceae), cv. ‘Broma’, on the basis of the analysis of issr markers. Acta Agrobot 65(3):35–42
Alonso A, Bull RD, Acedo C, Gillespie LJ (2014) Design of plant-specific PCR primers for the ETS region with enhanced specificity for tribe Bromeae and their application to other grasses (Poaceae). Botany 92:693–699
Paliwal R, Singh R, Singh AK, Kumar S, Kumar A, Singh MR (2013) Molecular characterization of Giloe (Tinospora cordifolia Willd. Miersex Hook. F. and Thoms.) accessions using start codon targeted (SCoT) markers. Int J Med Arom Plants 3(4):413–422
Wu JM, Li YR, Yang LT, Fang FX, Song HZ, Tang HQ, Wang M, Weng ML (2015) cDNA-SCoT: a novel rapid method for analysis of gene differential expression in sugarcane and other plants. Aust J Crop Sci 7(5):659–664
Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88:226–237
Xiong FQ, Zhong RC, Han ZQ, Jiang J, He LQ, Zhuang WJ, Tang RH (2011) Start codon targeted (SCoT) polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Torre-Bárcena JE, Kolokotronis SO, Lee EK, Stevenson DW, Brenner ED, Katari MS, Coruzzi GM, DeSalle R (2009) The impact of out-group choice and missing data on major seed plant phylogenetics using genome-wide EST data. PLoS ONE 4(6):e5764
Murry MG, Tompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
De La Rosa R, James C, Tobutt KR (2002) Isolation and characterization of polymorphic microsatellite in olive (Olea europaea L.) and their transferability to other genera in Oleaceae. Mol Ecol Notes 2:265–267
Pillay M, Hilu KW (1990) Chloroplast DNA variation in diploid and polyploid species of Bromus (Poaceae) subgenera Festucaria and Ceratochloa. Theor Appl Genet 80:326–332
Oja T, Jaaska V, Vislap V (2003) Breeding system, evolution and taxonomy of Bromus arvensis, B. japonicus and B. squarrosus (Poaceae). Plant Syst Evol 242:101–117
Acknowledgements
The authors would like to acknowledge all friends in the Department of Agronomy and Plant Breeding, Campus of Agriculture and Natural Resources, for their help. The financial support for this work was provided by the Razi University Grant No. 56747.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they did not have a conflict of interest.
Ethical approval
This study was approved by the Ethics Committee of Razi University, Iran.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Safari, H., Zebarjadi, A., Kahrizi, D. et al. The study of inter-specific relationships of Bromus genus based on SCoT and ISSR molecular markers. Mol Biol Rep 46, 5209–5223 (2019). https://doi.org/10.1007/s11033-019-04978-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04978-2