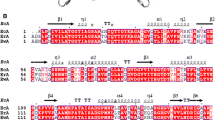
Abstract
Immunogenicity of therapeutic proteins is one of the main challenges in disease treatment. l-Asparaginase is an important enzyme in cancer treatment which sometimes leads to undesirable side effects such as immunogenic or allergic responses. Here, to decrease Erwinase (Erwinia chrysanthemil-Asparaginase) immunogenicity, which is the main drawback of the enzyme, firstly conformational B cell epitopes of Erwinase were predicted from three-dimensional structure by three different computational methods. A few residues were defined as candidates for reducing immunogenicity of the protein by point mutation. In addition to immunogenicity and hydrophobicity, stability and binding energy of mutants were also analyzed computationally. In order to evaluate the stability of the best mutant, molecular dynamics simulation was performed. Among mutants, H240A and Q239A presented significant reduction in immunogenicity. In contrast, the immunogenicity scores of D235A slightly decreased according to two servers. Binding affinity of substrate to the active site reduced significantly in K265A and E268A. The final results of molecular dynamics simulation indicated that H240A mutation has not changed the stability, flexibility, and the total structure of desired protein. Overall, point mutation can be used for reducing immunogenicity of therapeutic proteins, in this context, in silico approaches can be used to screen suitable mutants.








Similar content being viewed by others
References
Krishna M, Nadler SG (2016) Immunogenicity to biotherapeutics—the role of anti-drug immune complexes. Front Immunol 7:21. https://doi.org/10.3389/fimmu.2016.00021
Nechansky A, Kircheis R (2010) Immunogenicity of therapeutics: a matter of efficacy and safety. Expert Opin Drug Discov 5:1067–1079
Baker M, Reynolds HM, Lumicisi B, Bryson CJ (2010) Immunogenicity of protein therapeutics: the key causes, consequences and challenges. Self/nonself 1:314–322
Kintzing JR, Interrante MVF, Cochran JR (2016) Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends Pharmacol Sci 37(12):993–1008
Yari M, Ghoshoon BM, Vakili B, Ghasemi Y (2017) Therapeutic enzymes: applications and approaches to pharmacological improvement. Curr Pharm Biotechnol 18(7):531–540
Marshall SA, Lazar GA, Chirino AJ, Desjarlais JR (2003) Rational design and engineering of therapeutic proteins. Drug Discovery Today 8(5):212–221
Yao B, Zheng D, Liang S, Zhang C (2013) Conformational B-cell epitope prediction on antigen protein structures: a review of current algorithms and comparison with common binding site prediction methods. PLoS ONE 8(4):e62249
Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S (2015) An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform 53:405–414
Davids T, Schmidt M, Böttcher D, Bornscheuer UT (2013) Strategies for the discovery and engineering of enzymes for biocatalysis. Curr Opin Chem Biol 17(2):215–220
Zarei M, Nezafat N, Rahbar MR, Negahdaripour M, Sabetian S, Morowvat MH, Ghasemi Y (2018) Decreasing the immunogenicity of arginine deiminase enzyme via structure-based computational analysis. J Biomol Struct Dyn 1–14
Negahdaripour M, Nezafat N, Eslami M, Ghoshoon MB, Shoolian E, Najafipour S, Morowvat MH, Dehshahri A, Erfani N, Ghasemi Y (2018) Structural vaccinology considerations for in silico designing of a multi-epitope vaccine. Infect Genet Evol 58:96–109
Potocnakova L, Bhide M, Pulzova LB (2016) An introduction to B-Cell epitope mapping and in silico epitope prediction. J Immunol Res. https://doi.org/10.1155/2016/6760830
Liang S, Zheng D, Standley DM, Yao B, Zacharias M, Zhang C (2010) EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinform 11(1):381. https://doi.org/10.1186/1471-2105-11-381
Haste Andersen P, Nielsen M, Lund O (2006) Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci 15(11):2558–2567
Kringelum JV, Lundegaard C, Lund O, Nielsen M (2012) Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 8(12)
Ponomarenko J, Bui H-H, Li W, Fusseder N, Bourne PE, Sette A, Peters B (2008) ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform 9(1):514. https://doi.org/10.1186/1471-2105-9-514
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31(13):3381–3385
Colovos C, Yeates T (1993) ERRAT: an empirical atom-based method for validating protein structures. Protein Sci 2:1511–1519
Bowie JU, Luthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253(5016):164–170
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519. https://doi.org/10.1002/pro.5560020916
Lovell SC, Davis IW, Arendall WB, De Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50(3):437–450
Pandurangan AP, Ochoa-Montano B, Ascher DB, Blundell TL (2017) SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. https://doi.org/10.1093/nar/gkx439
Worth CL, Preissner R, Blundell TL (2011) SDM–a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 39:W215–W222. https://doi.org/10.1093/nar/gkr363
Lee B, Richards FM (1971) The interpretation of protein structures: estimation of static accessibility. J Mol Biol 55(3):379-IN374. https://doi.org/10.1016/0022-2836(71)90324-X
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157(1):105–132
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Hospital A, Goñi JR, Orozco M, Gelpí JL (2015) Molecular dynamics simulations: advances and applications. Adv Appl Bioinform Chem 8:37–47. https://doi.org/10.2147/AABC.S70333
Boehr DD, Nussinov R, Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5(11):789–796. https://doi.org/10.1038/nchembio.232
Hansson T, Oostenbrink C, van Gunsteren W (2002) Molecular dynamics simulations. Curr Opin Struct Biol 12(2):190–196
Abraham M, Van Der Spoel D, Lindahl E, Hess B (2014) The GROMACS development team. GROMACS User Manual Version 5(2):1–298
Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78(8):1950–1958
Nezafat N, Karimi Z, Eslami M, Mohkam M, Zandian S, Ghasemi Y (2016) Designing an efficient multi-epitope peptide vaccine against Vibrio cholerae via combined immunoinformatics and protein interaction based approaches. Comput Biol Chem 62:82–95
Nezafat N, Eslami M, Negahdaripour M, Rahbar MR, Ghasemi Y (2017) Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol Biosyst 13(4):699–713
Eslami M, Nezafat N, Khajeh S, Mostafavi-Pour Z, Bagheri Novir S, Negahdaripour M, Ghasemi Y, Razban V (2018) Deep analysis of N-cadherin/ADH-1 interaction: a computational survey. J Biomol Struct Dyn 1–57
Negahdaripour M, Golkar N, Hajighahramani N, Kianpour S, Nezafat N, Ghasemi Y (2017) Harnessing self-assembled peptide nanoparticles in epitope vaccine design. Biotechnol Adv 35(5):575–596
Imai K, Mitaku S (2005) Mechanisms of secondary structure breakers in soluble proteins. Biophysics 1:55–65. https://doi.org/10.2142/biophysics.1.55
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22(12):2577–2637
Kuriakose A, Chirmule N, Nair P (2016) Immunogenicity of biotherapeutics: causes and association with posttranslational modifications. J Immunol Res 2016:1298473. https://doi.org/10.1155/2016/1298473
Moola ZB, Scawen MD, Atkinson T, Nicholls DJ (1994) Erwinia chrysanthemi L-asparaginase: epitope mapping and production of antigenically modified enzymes. Biochem J 302(Pt 3):921–927
Van Regenmortel MH (2009) What is a B-cell epitope? Methods Mol Biol 524:3–20. https://doi.org/10.1007/978-1-59745-450-6_1
Ramya LN, Pulicherla KK (2015) Studies on deimmunization of antileukaemic l-asparaginase to have reduced clinical immunogenicity–an in silico approach. Pathol Oncol Res POR 21(4):909–920. https://doi.org/10.1007/s12253-015-9912-0
Meyer DL, Schultz J, Lin Y, Henry A, Sanderson J, Jackson JM, Goshorn S, Rees AR, Graves SS (2001) Reduced antibody response to streptavidin through site-directed mutagenesis. Protein Sci 10(3):491–503. https://doi.org/10.1110/ps.19901
Onda M, Nagata S, FitzGerald DJ, Beers R, Fisher RJ, Vincent JJ, Lee B, Nakamura M, Hwang J, Kreitman RJ, Hassan R, Pastan I (2006) Characterization of the B cell epitopes associated with a truncated form of Pseudomonas exotoxin (PE38) used to make immunotoxins for the treatment of cancer patients. J Immunology 177(12):8822–8834
Nagata S, Pastan I (2009) Removal of B cell epitopes as a practical approach for reducing the immunogenicity of foreign protein-based therapeutics. Adv Drug Deliv Rev 61(11):977–985. https://doi.org/10.1016/j.addr.2009.07.014
Collen D, Bernaerts R, Declerck P, De Cock F, Demarsin E, Jenné S, Laroche Y, Lijnen H, Silence K, Verstreken M (1996) Recombinant staphylokinase variants with altered immunoreactivity: I: construction and characterization. Circulation 94. https://doi.org/10.1161/01.CIR.94.2.197
Onda M, Beers R, Xiang L, Nagata S, Wang Q-C, Pastan I (2008) An immunotoxin with greatly reduced immunogenicity by identification and removal of B cell epitopes. Proc Natl Acad Sci USA 105(32):11311–11316
Kodama T, Hamakubo T, Doi H, Sugiyama A, Tsumoto K (2010) Hypo-immunogenic streptavidin and use thereof. International Patent Application WO/2010/095455 (August 26, 2010)
Acknowledgements
This study was supported by Grant No. 13435 from the Research Council of Shiraz University of Medical Sciences, Shiraz University of Medical Sciences, Shiraz, Iran.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yari, M., Eslami, M., Ghoshoon, M.B. et al. Decreasing the immunogenicity of Erwinia chrysanthemi asparaginase via protein engineering: computational approach. Mol Biol Rep 46, 4751–4761 (2019). https://doi.org/10.1007/s11033-019-04921-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04921-5