Abstract
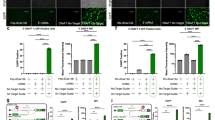
Pre-mRNA splicing is an essential step in gene expression, when introns are removed and exons joined by the complex of proteins called spliceosome. Correct splicing requires a precise exon/intron junction definition, which is determined by a consensual donor and acceptor splice site at the 5′ and 3′ end, respectively. An acceptor splice site (3′ss) consists of highly conserved AG nucleotides in positions E−2 and E−1. These nucleotides can appear in tandem, located 3 bp from each other. Then they are referred to as NAGNAG or tandem 3′ss, which can be alternatively spliced. NAG/TAG 3′ss motif abundance is extremely low and cannot be easily explained by just a nucleotide preference in this position. We tested artificial NAG/TAG motif’s potential negative effect on exon recognition using a minigene assay. Introducing the NAG/TAG motif into seven different exons revealed no general negative effect on exon recognition. The only observed effect was the partial use of the newly formed distal 3′ss. We can conclude that this motif’s extremely low preference in a natural 3′ss is not a consequence of the NAG/TAG motif’s negative effect on exon recognition, but more likely the result of other RNA processing aspects, such as an alternative 3′ss choice, decreased 3′ss strength, or incorporating an amber stop codon.




Similar content being viewed by others
References
Pan Q, Shai O, Lee LJ et al (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415. https://doi.org/10.1038/ng.259
Akerman M, Mandel-Gutfreund Y (2006) Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res 34:23–31. https://doi.org/10.1093/nar/gkj408
Dou Y, Fox-Walsh KL, Baldi PF, Hertel KJ (2006) Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA 12:2047–2056. https://doi.org/10.1261/rna.151106
Hiller M, Huse K, Szafranski K et al (2004) Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet 36:1255–1257. https://doi.org/10.1038/ng1469
Hiller M, Platzer M (2008) Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Genet 24:246–255. https://doi.org/10.1016/j.tig.2008.03.003
Yan X, Sablok G, Feng G et al (2015) nagnag: identification and quantification of NAGNAG alternative splicing using RNA-Seq data. FEBS Lett 589:1766–1770. https://doi.org/10.1016/j.febslet.2015.05.029
Tsai K-W, Tarn W-Y, Lin W-C (2007) Wobble splicing reveals the role of the branch point sequence-to-NAGNAG region in 3′ tandem splice site selection. Mol Cell Biol 27:5835–5848. https://doi.org/10.1128/MCB.00363-07
Hiller M, Szafranski K, Huse K et al (2008) Selection against tandem splice sites affecting structured protein regions. BMC Evol Biol 8:1–11. https://doi.org/10.1186/1471-2148-8-89
Grodecká L, Lockerová P, Ravčuková B et al (2014) Exon first nucleotide mutations in splicing: evaluation of in silico prediction tools. PLoS ONE. https://doi.org/10.1371/journal.pone.0089570
Hinzpeter A, Aissat A, Sondo E et al (2010) Alternative splicing at a NAGNAG acceptor site as a novel phenotype modifier. PLoS Genet 6:1–11. https://doi.org/10.1371/journal.pgen.1001153
Kent WJ, Sugnet CW, Furey TS et al (2002) The human genome browser at UCSC. Genome Res 12:996–1006. https://doi.org/10.1101/gr.229102
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394. https://doi.org/10.1089/1066527041410418
Zhang M (1998) Statistical features of human exons and their flanking regions. Hum Mol Genet 7:919–932. https://doi.org/10.1093/hmg/7.5.919
Shepard PJ, Choi E-A, Busch A, Hertel KJ (2011) Efficient internal exon recognition depends on near equal contributions from the 3′ and 5′ splice sites. Nucleic Acids Res 39:8928–8937. https://doi.org/10.1093/nar/gkr481
Hiller M, Huse K, Szafranski K et al (2006) Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am J Hum Genet 78:291–302. https://doi.org/10.1086/500151
Sinha R, Nikolajewa S, Szafranski K et al (2009) Accurate prediction of NAGNAG alternative splicing. Nucleic Acids Res 37:3569–3579. https://doi.org/10.1093/nar/gkp220
Akerman M, Mandel-Gutfreund Y (2007) Does distance matter? Variations in alternative 3′ splicing regulation. Nucleic Acids Res 35:5487–5498. https://doi.org/10.1093/nar/gkm603
Chern TM, Van Nimwegen E, Kai C et al (2006) A simple physical model predicts small exon length variations. PLoS Genet 2:606–613. https://doi.org/10.1371/journal.pgen.0020045
Aebi M, Hornig H, Padgett RA et al (1986) Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47:555–565. https://doi.org/10.1016/0092-8674(86)90620-3
Sorek R (2003) Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res 13:1631–1637. https://doi.org/10.1101/gr.1208803
Fu Y, Masuda A, Ito M et al (2011) AG-dependent 3′-splice sites are predisposed to aberrant splicing due to a mutation at the first nucleotide of an exon. Nucleic Acids Res 39:4396–4404. https://doi.org/10.1093/nar/gkr026
Bradley RK, Merkin J, Lambert NJ, Burge CB (2012) Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol. https://doi.org/10.1371/journal.pbio.1001229
Li L, Biswas K, Habib LA et al (2009) Functional redundancy of exon 12 of BRCA2 revealed by a comprehensive analysis of the c.6853A > G (p.I2285V) variant. Hum Mutat 30:1543–1550. https://doi.org/10.1002/humu.21101
Acknowledgements
This work was supported by the Ministry of Health of the Czech Republic, grant No. 16-34414A and by the Centre for Cardiovascular Surgery and Transplantation, grant No. 201606. Thanks should be given to Lucie Kopálková and Anna Žáková who provided technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hujová, P., Grodecká, L., Souček, P. et al. Impact of acceptor splice site NAGTAG motif on exon recognition. Mol Biol Rep 46, 2877–2884 (2019). https://doi.org/10.1007/s11033-019-04734-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04734-6