Abstract
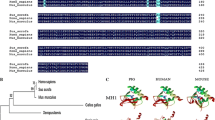
JHDM1A, a member of the JHDM (JmjC-domain-containing histone demethylase) family, plays an central role in gene silencing, cell cycle, cell growth and cancer development through histone H3K36 demethylation modification. Here reported the cloning, expression, chromosomal location and association analysis with growth traits of porcine JHDM1A gene. Sequence analysis showed that the porcine JHDM1A gene encodes 1,162 amino acids and contains JmjC, F-box, and CXXC zinc-finger domains, which coding sequence and deduced protein shares 91 and 99% similarity with human JHDM1A, respectively. Spatio-Temporal expression analysis indicated that the mRNA expression of porcine JHDM1A had significantly higher levels in the middle (65 days) and later (90 days) period’s embryo skeletal muscle than that of 33 days, and showed a ubiquitously expression but with the highest abundance in kidney, lung and liver of an adult pig. Radiation hybrid mapping and the following linkage mapping data indicate that JHDM1A maps to 2p17 region of pig chromosome 2 (SSC2). Allele frequency differences were detected in different pig breeds and an association study was performed with a SNP within 3′UTR. The results showed that there is a tendency for allele frequencies to differ between the fast growth breeds (Yorkshire) and slow growth pig breeds (Qingping pigs, Yushan Black pigs, Erhualian pigs and Dahuabai pigs). The association analysis using a Berkshire × Yorkshire F2 population indicated that the C224G polymorphism had a highly significant association with average daily gain on test (P < 0.01), a trend association with average back fat thickness (P < 0.07), and significant associations (P < 0.01) on percent of average drip loss, Fiber Type II Ratio, muscle shear force and average lactate content in μmol/g. This study provides the first evidence that JHDM1A is differentially expressed in porcine embryonic skeletal muscle and associated with meat growth and quality traits.




Similar content being viewed by others
References
Palacios D, Puri PL (2006) The epigenetic network regulating muscle development and regeneration. J Cell Physiol 207(1):1–11. doi:10.1002/jcp.20489
Tang Z, Li Y, Wan P, Li X, Zhao S, Liu B, Fan B, Zhu M, Yu M, Li K (2007) LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biol 8(6):R115. doi:10.1186/gb-2007-8-6-r115
Lin W, Dent SY (2006) Functions of histone-modifying enzymes in development. Curr Opin Genet Dev 16(2):137–142. doi:10.1016/j.gde.2006.02.002
Peng YB, Yerle M, Liu B (2009) Mapping and expression analyses during porcine foetal muscle development of 12 genes involved in histone modifications. Anim Genet 40(2):242–246. doi:10.1111/j.1365-2052.2008.01818.x
Pandorf CE, Haddad F, Wright C, Bodell PW, Baldwin KM (2009) Differential epigenetic modifications of histones at the myosin heavy chain genes in fast and slow skeletal muscle fibers and in response to muscle unloading. Am J Physiol Cell Physiol. doi:10.1152/ajpcell.00075.2009
McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA (2008) Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol 10(1):77–84. doi:10.1038/ncb1671
Klose RJ, Kallin EM, Zhang Y (2006) JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7(9):715–727. doi:10.1038/nrg1945
Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 18(21):2627–2638. doi:10.1101/gad.1241904
Mal AK (2006) Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J 25(14):3323–3334. doi:10.1038/sj.emboj.7601229
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439(7078):811–816. doi:10.1038/nature04433
Tian X, Fang J (2007) Current perspectives on histone demethylases. Acta Biochim Biophys Sin (Shanghai) 39(2):81–88. doi:10.1111/j.1745-7270.2007.00272.x
He J, Kallin EM, Tsukada Y, Zhang Y (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol 15(11):1169–1175. doi:10.1038/nsmb.1499
Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450(7167):309–313. doi:10.1038/nature06255
Frescas D, Guardavaccaro D, Kuchay SM, Kato H, Poleshko A, Basrur V, Elenitoba-Johnson KS, Katz RA, Pagano M (2008) KDM2A represses transcription of centromeric satellite repeats and maintains the heterochromatic state. Cell Cycle 7(22):3539–3547
Pfau R, Tzatsos A, Kampranis SC, Serebrennikova OB, Bear SE, Tsichlis PN (2008) Members of a family of JmjC domain-containing oncoproteins immortalize embryonic fibroblasts via a JmjC domain-dependent process. Proc Natl Acad Sci USA 105(6):1907–1912. doi:10.1073/pnas.0711865105
Peng YB, Guan HP, Fan B, Zhao SH, Xu XW, Li K, Zhu MJ, Yerle M, Liu B (2008) Molecular characterization and expression pattern of the porcine STARS, a striated muscle-specific expressed gene. Biochem Genet 46(9–10):644–651. doi:10.1007/s10528-008-9178-2
Malek M, Dekkers JC, Lee HK, Baas TJ, Rothschild MF (2001) A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. I. Growth and body composition. Mamm Genome 12(8):630–636. doi:10.1007/s003350020019
Green P, Falls K, Crooks S (1990) Documentation for CRIMAP, version 2.4. Washington University School of Medicine, St. Louis
Pothof J, van Haaften G, Thijssen K, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, Tijsterman M (2003) Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes Dev 17(4):443–448. doi:10.1101/gad.1060703
Xu X, Qiu H, Du ZQ, Fan B, Rothschild MF, Yuan F, Liu B (2010) Porcine CSRP3: polymorphism and association analyses with meat quality traits and comparative analyses with CSRP1 and CSRP2. Mol Biol Rep 37(1):451–459. doi:10.1007/s11033-009-9632-1
Rattink AP, Faivre M, Jungerius BJ, Groenen MA, Harlizius B (2001) A high-resolution comparative RH map of porcine chromosome (SSC) 2. Mamm Genome 12(5):366–370. doi:10.1007/s003350020012
Qiu H, Xu X, Fan B, Rothschild MF, Martin Y, Liu B (2010) Investigation of LDHA and COPB1 as candidate genes for muscle development in the MYOD1 region of pig chromosome 2. Mol Biol Rep 37(1):629–636. doi:10.1007/s11033-009-9882-y
Liu G, Jennen DG, Tholen E, Juengst H, Kleinwachter T, Holker M, Tesfaye D, Un G, Schreinemachers HJ, Murani E, Ponsuksili S, Kim JJ, Schellander K, Wimmers K (2007) A genome scan reveals QTL for growth, fatness, leanness and meat quality in a Duroc-Pietrain resource population. Anim Genet 38(3):241–252. doi:10.1111/j.1365-2052.2007.01592.x
Thomsen H, Lee HK, Rothschild MF, Malek M, Dekkers JC (2004) Characterization of quantitative trait loci for growth and meat quality in a cross between commercial breeds of swine. J Anim Sci 82(8):2213–2228
de Koning DJ, Harlizius B, Rattink AP, Groenen MA, Brascamp EW, van Arendonk JA (2001) Detection and characterization of quantitative trait loci for meat quality traits in pigs. J Anim Sci 79(11):2812–2819
Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25(6):801–812. doi:10.1016/j.molcel.2007.03.001
Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG (2006) Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J 25(14):3422–3431. doi:10.1038/sj.emboj.7601215
Acknowledgments
This study was supported by the National High Science and Technology Foundation of China (2007AA10Z168) and the National Natural Science Foundation of China (30771536, 30900816).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Peng, Yb., Fan, B., Han, Xl. et al. Molecular characterization of the porcine JHDM1A gene associated with average daily gain: evaluation its role in skeletal muscle development and growth. Mol Biol Rep 38, 4697–4704 (2011). https://doi.org/10.1007/s11033-010-0604-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0604-2