Abstract
Broomrape (Orobanche cumana) is a parasitic weed that causes substantial yield losses in sunflower. In this study, four biparental genetic populations comprised of between 96 and 150 F3 families were phenotyped for resistance to broomrape race G. Bulk segregant analyses (BSA) combined with genotyping-by-sequencing (GBS) technology was used to verify previously identified genes and map new resistance QTLs. Resistance had a polygenic basis, and numerous QTLs were found in all mapping populations. Contributing components to resistance that were common to all populations mapped to two major QTLs on chromosome 3, which were designated or3.1 and or3.2. QTL or3.1 was positioned in a genomic region where a previous broomrape resistance gene Or5 has been mapped, while QTL or3.2 was identified for the first time in the lower region of the same chromosome. Following the identification of major QTLs for resistance using the BSA-seq approach, all plants from three F2 populations were genotyped using newly developed CAPS markers nearest to the QTL peak for or3.2, which confirmed the association of these markers with broomrape race G resistance. The results of this study will bring us a step closer to understanding the mechanisms underlying resistance of sunflower to highly virulent broomrape races, and the molecular markers developed herein will be highly useful for sunflower breeding programs.


Similar content being viewed by others
References
Akhtouch B, del Moral L, Leon A, Velasco L, Fernández-Martínez JM, Pérez-Vich B (2016) Genetic study of recessive broomrape resistance in sunflower. Euphytica 209:419–428
Akhtouch B, Muñoz-Ruz J, Melero-Vara J, Fernández-Martínez J, Domínguez J (2002) Inheritance of resistance to race F of broomrape in sunflower lines of different origins. Plant Breed 121:266–268
Alonso L, Lopez G, MI RO, Sallago F (1996) New highly virulent sunflower broomrape (Orobanche cernua Loefl.) pathotypes in Spain. In: Congresos y Jornadas-Junta de Andalucia (Espana). no. 36/96
Amri M, Abbes Z, Youssef SB, Bouhadida M, Salah HB, Kharrat M (2012) Detection of the parasitic plant, Orobanche cumana on sunflower (Helianthus annuus L.) in Tunisia. Afr J Biotechnol 11:4163–4167
Antonova TS (2014) The history of interconnected evolution of Orobanche cumana Wallr. and sunflower in the Russian Federation and Kazakhstan. Helia 37:215–225
CABI (2018) Orobanche cumana (sunflower broomrape). In: Invasive species compendium. Wallingford, UK: CAB International. www.cabi.org/isc. Accessed 14 October 2018
Caplan J, Padmanabhan M, Dinesh-Kumar SP (2008) Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3:126–135
Chater AO, Webb DA (1972) Orobanche. In: Tutin TG, Heywood VH, Burgess NA, Morre DM, Valentine, DH, Walters SM, Webb DM (eds) Flora Europaea, vol 3, Diapensiaceae to Myoporaceae, University Press, Cambridge, pp 286–293
Christov M, Piskov A, Encheva J, Valkova D, Drumeva M, Nenova N et al (2009) Developing sunflower hybrid cultivars with increased productivity, resistant to disease and broomrape using classical and biotechnological methods. Science-technical bulletin. Institute for Oilseed Crops. UOSC 74–87
Danial DL, Stubbs RW, Parlevliet JE (1994) Evolution of virulence patterns in yellow rust races and its implications for breeding for resistance in wheat in Kenya. Euphytica 80:165–170
Fernández-Aparicio M, Sillero JC, Rubiales D (2009) Resistance to broomrape species (Orobanche spp.) in common vetch (Vicia sativa L.). Crop Prot 28:7–12
Fernández-Martínez J, Melero-Vara J, Muñoz-Ruz J, Ruso J, Domínguez J (2000) Selection of wild and cultivated sunflower for resistance to a new broomrape race that overcomes resistance of the Or5 gene. Crop Sci 40:550–555
Fernández-Martínez J, Pérez-Vich B, Akhtouch B, Velasco L (2004) Registration of four sunflower germplasms resistant to race F of broomrape. Crop Sci 44:1033
Fernández-Martínez J, Velasco L, Pérez-Vich B (2012) Progress in research on breeding for resistance to sunflower broomrape. Helia 35:47–56
González-Torres R, Jiménez-Díaz R, Melero-Vara J (1982) Distribution and virulence of Orobanche cernua in sunflower crops in Spain. J Phytopathol 104:78–89
Grenz J, Manschadi A, Uygur F, Sauerborn J (2005) Effects of environment and sowing date on the competition between faba bean (Vicia faba) and the parasitic weed Orobanche crenata. Field Crop Res 93:300–313
Gressel J, Hanafi A, Head G, Marasas W, Obilana AB, Ochanda J, Souissi T, Tzotzos G (2004) Major heretofore intractable biotic constraints to African food security that may be amenable to novel biotechnological solutions. Crop Prot 23:661–689
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363
Imerovski I, Dimitrijević A, Miladinović D, Dedić B, Jocić S, Kočiš Tubić N et al (2016) Mapping of a new gene for resistance to broomrape races higher than F. Euphytica 209:281–289
Jan C, Fernández-Martínez J, Ruso J, Muñoz-Ruz J (2002) Registration of four sunflower germplasms with resistance to Orobanche cumana race F. Crop Sci 42:2217
Jocić S, Cvejić S, Jocković M, Hladni N, Dedić B, Imerovski I, Miladinović D, Miklič V (2016) Screening for resistance to highly virulent races of sunflower broomrape (Orobanche cumana). Proc. 19th Int. Sunflower Conf., Edirne, Turkey, 29 May-3 June, pp 534
Jocković M, Jocić S, Cvejić S, Miladinović D, Dedić B, Terzić S, Marjanović-Jeromela A, Miklič V (2018) Helianthus species as a sources for broomrape resistance. Proc. 4th Int. Symp. Broomrape in Sunflower, Bucharest, Romania, 2–4 July, pp 178–186
Joița M, Fernández-Martínez J, Sava E, Raranciuc S (2009) Broomrape (Orobanche cumana Wallr.), the most important parasite in sunflower. Analele Institutului Național de Cercetare-Dezvoltare Agricolă Fundulea 77:49–56
Kaya Y, Evci G, Pekcan V, Gucer T (2004) Determining new broomrape-infested areas, resistant lines and hybrids in Trakya region of Turkey. Helia 27:211–218
Kleinhofs A, Brueggeman R, Nirmala J, Zhang L, Mirlohi A, Druka A, Rostoks N, Steffenson BJ (2009) Barley stem rust resistance genes: structure and function. The Plant Genome 2:109–120
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Lambel S, Lanini B, Vivoda E, Fauve J, Wechter WP, Harris-Shultz KR et al (2014) A major QTL associated with Fusarium oxysporum race 1 resistance identified in genetic populations derived from closely related watermelon lines using selective genotyping and genotyping-by-sequencing for SNP discovery. Theor Appl Genet 127:2105–2115
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li J, Timko MP (2009) Gene-for-gene resistance in Striga-cowpea associations. Science 325:1094–1094
Li Z-K, Luo L, Mei H, Paterson A, Zhao X, Zhong D et al (1999) A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol Gen Genet 261:58–63
Li Z, Pinson SR, Park WD, Paterson AH, Stansel JW (1997) Epistasis for three grain yield components in rice (Oryxa sativa L.). Genetics 145:453–465
Louarn J, Boniface M-C, Pouilly N, Velasco L, Pérez-Vich B, Vincourt P et al (2016) Sunflower resistance to broomrape (Orobanche cumana) is controlled by specific QTLs for different parasitism stages. Front Plant Sci 7:590
Magwene PM, Willis JH, Kelly JK (2011) The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput Biol 7:e1002255
Manschadi A, Kroschel J, Sauerborn J (1996) Dry matter production and partitioning in the host-parasite association Vicia faba-Orobanche crenata. Angew Bot 70:224–229
Mansfeld BN, Grumet R (2018) QTLseqr: an R package for bulk segregant analysis with next-generation sequencing. The Plant Genome 11(2):1–5. https://doi.org/10.3835/plantgenome2018.01.0006
Martín-Sanz A, Malek J, Fernández-Martínez JM, Pérez-Vich B, Velasco L (2016) Increased virulence in sunflower broomrape (Orobanche cumana Wallr.) populations from southern Spain is associated with greater genetic diversity. Front Plant Sci 7:589
Maširević S, Medić-Pap S, Škorić D (2012) Is there appearance of new broomrape race in Serbia. Proc. 18th Int. Conf. in Sunflower, Mar del Plata, Argentina, 2–4 February, pp 1048–1051
Melero-Vara J, Dominguez J, Fernandez-Martinez J (1989) Evaluation of differential lines and a collection of sunflower parental lines for resistance to broomrape (Orobanche cernua) in Spain. Plant Breed 102:322–326
Molinero-Ruiz L, Delavault P, Pérez-Vich B, Păcureanu-Joita M, Bulos M, Altieri E et al (2015) History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed: a review. Span J AgricRes 13:10–11
Molinero-Ruiz M, Perez-Vich B, Pineda-Martos R, Melero-Vara J (2008) Indigenous highly virulent accessions of the sunflower root parasitic weed Orobanche cumana. Weed Res 48:169–178
Nass H, Pedersen W, MacKenzie D, Nelson R (1981) The residual effects of some “defeated” powdery mildew resistance genes in isolines of winter wheat [Erysiphe graminis f. sp. tritici]. Phytopathology (USA)
Natali L, Cossu RM, Barghini E, Giordani T, Buti M, Mascagni F, Morgante M, Gill N, Kane NC, Rieseberg L, Cavallini A (2013) The repetitive component of the sunflower genome as shown by different procedures for assembling next generation sequencing reads. BMC Genomics 14(1):686
Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P (2018) Navigating complexity to breed disease-resistant crops. Nat Rev Genet 19(21)
Owens GL, Baute GJ, Rieseberg LH (2016) Revisiting a classic case of introgression: hybridization and gene flow in Californian sunflowers. Mol Ecol25 25:2630–2643
Păcureanu JM, Raranciuc S, Sava E, Stanciu D, Nastase D (2009) Virulence and aggressiveness of sunflower broomrape (Orobanche cumana Wallr.) populations in Romania. Helia 32:111–117
Parker C (2013) The parasitic weeds of the Orobanchaceae. In: Joel DM, Gressel J, Musselman LJ (eds) Parasitic Orobanchaceae. Springer, New York, pp 313–344
Parlevliet JE (2002) Durability of resistance against fungal, bacterial and viral pathogens; present situation. Euphytica 124:147–156
Pérez-Vich B, Akhtouch B, Knapp S, Leon A, Velasco L, Fernández-Martínez J et al (2004a) Quantitative trait loci for broomrape (Orobanche cumana Wallr.) resistance in sunflower. Theor Appl Genet 109:92–102
Pérez-Vich B, Akhtouch B, Muñoz-Ruz J, Fernández-Martínez J, Jan C (2002) Inheritance of resistance to a highly virulent race F of Orobanche cumana Wallr. In a sunflower line derived from interspecific amphiploids. Helia 25:137–144
Pérez-Vich B, Aktouch B, Mateos A, Velasco L, Jan C, Fernández J et al (2004b) Dominance relationships for genes conferring resistance to broomrape (Orobanche cumana Wallr.) in sunflower. Helia 27:183–192
Permingeat HR, Romagnoli MV, Sesma JI, Vallejos RH (1998) A simple method for isolating DNA of high yield and quality from cotton (shape Gossypium hirsutum L.) leaves. Plant Mol Biol Report 16:89–89
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7(2):e32253. https://doi.org/10.1371/journal.pone.0032253
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ, pp 365–386
Schneeberger K, Weigel D (2011) Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci 16:282–288
Sedlazeck FJ, Rescheneder P, Von Haeseler A (2013) NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29:2790–2791
Shindrova P (1994) Distribution and race composition of Orobanche cumana Wallr. in Bulgaria. In: Biology and management of Orobanche. Proc.3rd Int. Workshop Orobanche and related Striga research, Royal Tropical Institute, Amsterdam, Netherlands, 8–12 November 1993, pp 142–145
Škorić D, Păcureanu M (2010) Sunflower breeding for resistance to broomrape (Orobanche cumana Wallr.). Proc. Int. Symp. Sunflower Breeding on Resistance to Diseases, Krasnodar, Russia,22–24 June, pp 19–30
Stewart HE, Bradshaw JE, Pande B (2003) The effect of the presence of R‐genes for resistance to late blight (Phytophthora infestans) of potato (Solanum tuberosum) on the underlying level of field resistance. Plant Pathol 52:193–198
Sukno S, Melero-Vara J, Fernández-Martínez J (1999) Inheritance of resistance to Orobanche cernua Loefl. in six sunflower lines. Crop Sci 39:674–678
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183
Tameling WI, Joosten MH (2007) The diverse roles of NB-LRR proteins in plants. Physiol Mol Plant Pathol 71(4–6):126–134
Tang S, Heesacker A, Kishore VK, Fernandez A, Sadik ES, Cole G, Knapp SJ (2003) Genetic mapping of the Or 5 gene for resistance to Orobanche race E in sunflower. Crop Sci 43:1021–1028
Van der Auwera G, Carneiro M, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al (2013) From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 43:1–33
Velasco L, Pérez-Vich B, Yassein AA, Jan CC, Fernández-Martínez JM (2012) Inheritance of resistance to sunflower broomrape (Orobanche cumana Wallr) in an interspecific cross between Helianthus annuus and Helianthus debilis subsp tardiflorus. Plant Breed 131:220–221
Velasco L, Pérez-Vich B, Jan C, Fernández-Martínez J (2007) Inheritance of resistance to broomrape (Orobanche cumana Wallr.) race F in a sunflower line derived from wild sunflower species. Plant Breed 126:67–71
Vrânceanu A, Păcureanu M (1995) Evaluation of an international set of sunflower hybrids in relation to broomrape (Orobanche cumana Wallr.) resistance. Rom Agric Res 19–24
Vrânceanu A, Tudor V, Stoenescu F, Pirvu N (1980) Virulence groups of Orobanche cumana Wallr. differential hosts and resistance sources and genes in sunflower. Proc. 9th Int. Sunflower Conf., Torremolinos, Spain,8–13 June, pp 74–82
Wetterstrand KA (2018) DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP).www.genome.gov/sequencingcostsdata. Accessed 23 March 2018
Win KT, Vegas J, Zhang C, Song K, Lee S (2017) QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor Appl Genet 130:199–211
Wright K (2018) Graphical Gems in the agridat Package. https://cran.r-project.org/web/packages/agridat/vignettes/agridat_examples.html. Accessed 6 April 2018
Yan W, Cornelius PL, Crossa J, Hunt L (2001) Two types of GGE biplots for analyzing multi-environment trial data. Crop Sci 41:656–663
Yan W, Hunt L, Sheng Q, Szlavnics Z (2000) Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci 40:597–605
Yan W, Kang MS (2002) GGE biplot analysis: a graphical tool for breeders, geneticists, and agronomists, CRC press, Boca Raton, FL, USA
Yang Z, Huang D, Tang W, Zheng Y, Liang K, Cutler AJ, Wu W (2013) Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS One 8:e68433
Yoder JI, Scholes JD (2010) Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr Opin Plant Biol 13:478–484
Zhang J, Li X, Jiang G, Xu Y, He Y (2006) Pyramiding of Xa7 and Xa21 for the improvement of disease resistance to bacterial blight in hybrid rice. Plant Breed 125:600–605
Funding
This work was in part supported by project TR31025 financed by the Ministry of Education, Science and Development, Republic of Serbia and funding from Genome BC and Genome Canada to LHR.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Table S1
Description of the environments (combination of location and season) of the trials for the multi-environment study (DOCX 21 kb)
Table S2
Relative broomrape incidence (in %) of four selected sunflower inbred lines, their F1s and differential line set in the eight different environments. Relative values expressed as percentage of the susceptible control Labud (DOCX 25 kb)
Table S3
Accession numbers of biosamples deposited in SRA (DOCX 20 kb)
Table S4
Initial dataset size and number of SNPs per population after filtering within “QTLseqr” (DOCX 20 kb)
Table S5
CAPS markers developed from SNPs obtained by genotype-by-sequencing (DOCX 20 kb)
Fig. S1
(a) Testing sites used for germplasm screening in year 2011 and for multi-environmental trials on selected lines during 2016 and 2017. (b) Within the selected sites, broomrape incidence rate on the differential line LC1093 (carrying gene Or6) is shown with the pie-charts. The size of the pie-charts is scaled to reflect the infestation rate on LC1093 (i.e. number of broomrape plant on the host plant). The map was generated using averages from data collected during 2011–2016 (JPG 696 kb)
Fig. S2
Frequency distribution of broomrape resistance within 300 sunflower lines evaluated in the field in Serbia (a), Spain (b), Constanta, Romania (c) and Tulcea, Romania(d) during summer of 2011. (e) Venn diagram representation of number of resistant lines in the four environments. Lines with a score of 0 or 1 were considered as resistant, whereas all other lines were considered susceptible. Seven lines were resistant in all four environments (PNG 1005 kb)
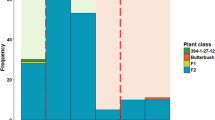
Fig. S3
Frequency distribution of broomrape resistance scores of the F3 families in the populations (a) HA-267/OD-DI-82, (b) LIV-10/HA-26-PR, (c) LIV-17/HA-26-PR and (d) AB-VL-8/L-OS-1. (PDF 1650 kb)
Rights and permissions
About this article
Cite this article
Imerovski, I., Dedić, B., Cvejić, S. et al. BSA-seq mapping reveals major QTL for broomrape resistance in four sunflower lines. Mol Breeding 39, 41 (2019). https://doi.org/10.1007/s11032-019-0948-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0948-9