Abstract
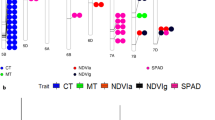
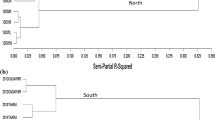
Winter survival ability is important for autumn sown winter wheat (Triticum aestivum L.) in regions with cold winters. Wheat vernalization and photoperiod genes influence adaptation by regulating the timing of the transition from vegetative to reproductive growth to protect the floral meristem from cold temperatures. We evaluated winter injury of 287 genotypes from the Facultative and Winter Wheat Observation Nursery (FAWWON) in six field environments over 3 years (2014 to 2016) in Colorado. Entries were genotyped using single-nucleotide polymorphisms (SNPs) obtained by genotyping by sequencing (GBS) and at known vernalization (Vrn-A1, Vrn-B1, and Vrn-D1) and photoperiod (Ppd-B1 and Ppd-D1) loci using Kompetitive Allele Specific PCR (KASP) assays. Winter injury was observed and visually scored in five of the six environments. Mean GS prediction accuracies across the five environments, obtained through ridge regression best linear unbiased prediction (RR-BLUP) using 23,269 SNPs alone as random effects, ranged from 0.26 ± 0.01 to 0.74 ± 0.00. Incorporation of alleles at Vrn-A1, Vrn-B1, and Vrn-D1 loci as fixed effects in the GS models together with GBS markers as random effects provided the highest prediction accuracy with mean GS accuracies ranging from 0.34 ± 0.01 to 0.78 ± 0.00 across the five environments. Genomic selection models incorporating photoperiod alleles as fixed effects rarely improved GS prediction accuracy of winter injury. Genomic selection models that incorporate both major and minor genetic factors that influence low-temperature tolerance improved the model predictions for identifying genotypes that are best adapted to regions where cold winter temperatures are an important production constraint.



Similar content being viewed by others
Data availability
The SNP genotype and phenotypic datasets generated during and/or analyzed during the current study are freely available from the corresponding author upon request.
References
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115:721–733. https://doi.org/10.1007/s00122-007-0603-4
Bernardo R (2008) Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci 48:1649–1664. https://doi.org/10.2135/cropsci2008.03.0131
Bernardo R (2014) Genomewide selection when major genes are known. Crop Sci 54:68–75. https://doi.org/10.2135/cropsci2013.05.0315
Braun H-J, Sãulescu NN (2002) Breeding winter and facultative wheat. In: Curtis BC, Rajaram S, Gómez MH (eds) Bread wheat: improvement and production. Food and Agriculture Organization of the United Nations, Rome, pp 217–226
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype × environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719. https://doi.org/10.2135/cropsci2011.06.0299
Busch R, Elsayed F, Heiner R (1984) Effect of daylength insensitivity on agronomic traits and grain protein in hard red spring wheat. Crop Sci 24:1106–1109. https://doi.org/10.2135/cropsci1984.0011183X002400060023x
Butler D (2009) asreml: asreml() fits the linear mixed model. R package version 3.0. https://www.vsni.co.uk/downloads/asreml-r/. Accessed 5 January 2018
Chen Y, Carver BF, Wang S, Zhang F, Yan L (2009) Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor Appl Genet 118:881–889. https://doi.org/10.1007/s00122-008-0946-5
Cobb JN, DeClerck G, Greenberg A, Clark R, McCouch S (2013) Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor Appl Genet 126:867–887. https://doi.org/10.1007/s00122-013-2066-0
Cockram J, Jones H, Leigh FJ, O'Sullivan D, Powell W, Laurie DA, Greenland AJ (2007) Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot 58:1231–1244. https://doi.org/10.1093/jxb/erm042
Coombes NE (2009) DiGGer design search tool in R. http://nswdpibiom.org/austatgen/software. Accessed 5 January 2018
Crosatti C, Mare C, Massucotelli E, Belloni S, Barilli S, Bassi R, Dubcovsky J, Galiba G, Stanca A, Cattivelli L (2003) Genetic analysis of the expression of the cold-regulated gene cor14b: a way toward the identification of components of cold response signal transduction in Triticeae. Can. J. Bot. 81: 1162–1167.
Crossa J, de Los Campos G, Pérez P, Gianola D, Burgueño J, Araus JL, Makumbi D, Singh RP, Dreisigacker S, Yan J (2010) Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186:713–724. https://doi.org/10.1534/genetics.110.118521
Daetwyler HD, Bansal UK, Bariana HS, Hayden MJ, Hayes BJ (2014) Genomic prediction for rust resistance in diverse wheat landraces. Theor Appl Genet 127:1795–1803. https://doi.org/10.1007/s00122-014-2341-8
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B Methodol 39:1–38
Dhillon T, Pearce SP, Stockinger EJ, Distelfeld A, Li C, Knox AK, Vashegyi I, Vágújfalvi A, Galiba F, Dubcovsky J (2010) Regulation of freezing tolerance and flowering in temperate cereals: the VRN-1 connection. Plant Physiol 153:1846–1858
Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA (2012) Copy number variation affecting the photoperiod-B1 and vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7:e33234. https://doi.org/10.1371/journal.pone.0033234
Distelfeld A, Li C, Dubcovsky J (2009) Regulation of flowering in temperate cereals. Curr Opin Plant Biol 12:178–184. https://doi.org/10.1016/j.pbi.2008.12.010
Eagles H, Cane K, Kuchel H, Hollamby G, Vallance N, Eastwood R, Gororo N, Martin P (2010) Photoperiod and vernalization gene effects in southern Australian wheat. Crop Pasture Sci 61:721–730. https://doi.org/10.1071/CP10121
Eagles HA, Cane K, Trevaskis B (2011) Veery wheats carry an allele of Vrn-A1 that has implications for freezing tolerance in winter wheats. Plant Breed 130:413–418. https://doi.org/10.1111/j.1439-0523.2011.01856.x
Endelman JB (2011) Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 4:250–255. https://doi.org/10.3835/plantgenome2011.08.0024
Fowler DB (2012) Wheat production in the high winter stress climate of the great plains of North America—an experiment in crop adaptation. Crop Sci 52:11–20. https://doi.org/10.2135/cropsci2011.05.0279
Fowler DB, Gusta L, Tyler N (1981) Selection for winterhardiness in wheat. III. Screening methods. Crop Sci 21:896–901. https://doi.org/10.2135/cropsci1981.0011183X002100060023x
Fowler DB, Limin A, Wang S-Y, Ward R (1996) Relationship between low-temperature tolerance and vernalization response in wheat and rye. Can J Plant Sci 76:37–42. https://doi.org/10.4141/cjps96-007
Fu D, Szűcs P, Yan L, Helguera M, Skinner JS, Von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Gen Genomics 273:54–65. https://doi.org/10.1007/s00438-004-1095-4
Galiba G, Vágújfalvi A, Li C, Soltész A, Dubcovsky J (2009) Regulatory genes involved in the determination of frost tolerance in temperate cereals. Plant Sci 176:12–19. https://doi.org/10.1016/j.plantsci.2008.09.016
Gardner F, Barnett R (1990) Vernalization of wheat cultivars and a triticale. Crop Sci 30:166–169. https://doi.org/10.2135/cropsci1990.0011183X003000010036x
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9(2):e90346. https://doi.org/10.1371/journal.pone.0090346
Grogan SM, Brown-Guedira G, Haley SD, McMaster GS, Reid SD, Smith J, Byrne PF (2016) Allelic variation in developmental genes and effects on winter wheat heading date in the US Great Plains. PLoS One 11:e0152852. https://doi.org/10.1371/journal.pone.0152852
Gusta LV, Wisniewski M (2013) Understanding plant cold hardiness: an opinion. Physiol Plant 147:4–14. https://doi.org/10.1111/j.1399-3054.2012.01611.x
Haley SD, Johnson JJ, Peairs FB, Stromberger JA, Hudson EE, Seifert SA, Kottke RA, Valdez VA, Rudolph JB, Bai G, Chen X, Bowden RL, Jin Y, Kolmer JA, Chen M-S, Seabourn BW (2012) Registration of ‘Byrd’ wheat. J Plant Reg 6:302–305. https://doi.org/10.3198/jpr2011.12.0672crc
Heffner EL, Sorrells ME, Jannink JL (2009) Genomic selection for crop improvement. Crop Sci 49:1–12. https://doi.org/10.2135/cropsci2008.08.0512
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. https://doi.org/10.1126/science.1251788
Iwaki K, Haruna S, Niwa T, Kato K (2001) Adaptation and ecological differentiation in wheat with special reference to geographical variation of growth habit and Vrn genotype. Plant Breed 120:107–114. https://doi.org/10.1046/j.1439-0523.2001.00574.x
Jannink J-L, Lorenz AJ, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Brief Funct Genomics 9:166–177. https://doi.org/10.1093/bfgp/elq001
John JA, Williams ER (1995) Cyclic and computer generated designs, 2nd edn. St. Edmundsbury Press, Bury St. Edmunds ISBN 9780412575808
Kamran A, Iqbal M, Spaner D (2014) Flowering time in wheat (Triticum aestivum L.): a key factor for global adaptability. Euphytica 197:1–26. https://doi.org/10.1007/s10681-014-1075-7
Kiss T, Balla K, Veisz O, Láng L, Bedő Z, Griffiths S, Isaac P, Karsai I (2014) Allele frequencies in the VRN-A1, VRN-B1 and VRN-D1 vernalization response and PPD-B1 and PPD-D1 photoperiod sensitivity genes, and their effects on heading in a diverse set of wheat cultivars (Triticum aestivum L.). Mol Breed 34:297–310. https://doi.org/10.1007/s11032-014-0034-2
Law C, Sutka J, Worland A (1978) A genetic study of day-length response in wheat. Heredity 41:185–191. https://doi.org/10.1038/hdy.1978.87
Limin AE, Fowler DB (2006) Low-temperature tolerance and genetic potential in wheat (Triticum aestivum L.): response to photoperiod, vernalization, and plant development. Planta 224:360–366. https://doi.org/10.1007/s00425-006-0219-y
Lin F, Xue SL, Tian DG, Li CJ, Cao Y, Zhang ZZ, Shang CQ, Ma ZQ (2008) Mapping chromosomal regions affecting flowering time in a spring wheat RIL population. Euphytica 164:769–777. https://doi.org/10.1007/s10681-008-9724-3
Lorenz AJ, Smith KP (2015) Adding genetically distant individuals to training populations reduces genomic prediction accuracy in barley. Crop Sci 55:2657–2667. https://doi.org/10.2135/cropsci2014.12.0827
Mahfoozi S, Limin A, Hayes P, Hucl P, Fowler DB (2000) Influence of photoperiod response on the expression of cold hardiness in wheat and barley. Can J Plant Sci 80:721–724. https://doi.org/10.4141/P00-031
Marshall L, Busch R, Cholick F, Edwards I, Frohberg R (1989) Agronomic performance of spring wheat isolines differing for daylength response. Crop Sci 29:752–757. https://doi.org/10.2135/cropsci1989.0011183X002900030043x
Meuwissen T, Hayes B, Goddard M (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Moore JK, Manmathan HK, Anderson VA, Poland JA, Morris CF, Haley SD (2017) Improving genomic prediction for pre-harvest sprouting tolerance in wheat by weighting large-effect quantitative trait loci. Crop Sci 57:1315–1324. https://doi.org/10.2135/cropsci2016.06.0453
Morgounov A, Braun H-J, Ketata H, Paroda R (2005) International cooperation for winter wheat improvement in central Asia: results and perspectives. Turk J Agric For 29:137–142
Morgounov A, Tufan HA, Sharma R, Akin B, Bagçi A, Braun H-J, Kaya Y, Keser M, Payne TS, Sonder K (2012) Global incidence of wheat rusts and powdery mildew during 1969–2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur J Plant Pathol 132:323–340. https://doi.org/10.1007/s10658-011-9879-y
Paulsen GM, Shroyer JP (2008) The early history of wheat improvement in the Great Plains. Agron J 100:S-70–S-78. https://doi.org/10.2134/agronj2006.0355c
Peterson C, Pfeiffer W (1989) International winter wheat evaluation: relationships among test sites based on cultivar performance. Crop Sci 29:276–282. https://doi.org/10.2135/cropsci1989.0011183X002900020008x
Piepho HP, Möring J, Melchinger AE, Büchse A (2008) BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161:209–228. https://doi.org/10.1007/s10681-007-9449-8
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253. https://doi.org/10.1371/journal.pone.0032253
Pugsley A (1966) The photoperiodic sensitivity of some spring wheats with special reference to the variety Thatcher. Aust J Agric Res 17:591–599
Pugsley A (1971) A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res Sci 22:21–31. https://doi.org/10.1071/AR9710021
Pugsley A (1972) Additional genes inhibiting winter habit in wheat. Euphytica 21:547–552. https://doi.org/10.1007/BF00039355
Randhawa HS, Asif M, Pozniak C, Clarke JM, Graf RJ, Fox SL, Humphreys DG, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Hucl P, Spaner, D (2013) Application of molecular markers to wheat breeding in Canada. Plant Breed. 132: 458–471. https://doi.org/10.1111/pbr.12057
R Development Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. Accessed 5 January 2018
Rajaram S, Van Ginkel M, Fischer RA (1993) CIMMYT’s wheat breeding mega-environments (ME). p. 1101-1106. In Proc. of the 8th Int. Wheat Genetic Symp, Beijing 20–25 July 1993. Institute of Genetics, Chinese Academy of Sciences, Beijing
Rapacz M, Woźniczka A (2009) A selection tool for freezing tolerance in common wheat using the fast chlorophyll a fluorescence transient. Plant Breed 128:227–234. https://doi.org/10.1111/j.1439-0523.2008.01566.x
Rousset M, Bonnin I, Remoué C, Falque M, Rhoné B, Veyrieras J-B, Madur D, Murigneux A, Balfourier F, Le Gouis J (2011) Deciphering the genetics of flowering time by an association study on candidate genes in bread wheat (Triticum aestivum L.). Theor Appl Genet 123:907–926. https://doi.org/10.1007/s00122-011-1636-2
Santra DK, Santra M, Allan R, Garland-Campbell K, Kidwell K (2009) Genetic and molecular characterization of vernalization genes Vrn-A1, Vrn-B1, and Vrn-D1 in spring wheat germplasm from the Pacific northwest region of the USA. Plant Breed 128:576–584. https://doi.org/10.1111/j.1439-0523.2009.01681.x
Scarth R, Law C (1984) The control of the day-length response in wheat by the group 2 chromosomes. Z Pflanzenzüchtung 92:140–150
Sharma R, Morgounov A, Akin B, Bespalova L, Láng L, Litvinenko M, Mustatea P, Ozturk I, Postolatiy A, Rajaram S (2014) Winter wheat eastern European regional yield trial: identification of superior genotypes and characterization of environments. Crop Sci 54:2469–2480. https://doi.org/10.2135/cropsci2014.01.0028
Stushnoff C, Fowler B, Brule-Babel A (1984) Breeding and selection for resistance to low temperature. In: Vose PB (ed) Plant breeding-a contemporary basis. Pergamon Press, Oxford, pp 115–136 ISBN: 9780444597601
Sutka J (1994) Genetic control of frost tolerance in wheat (Triticum aestivum L.). Euphytica 77:277–282. https://doi.org/10.1007/BF02262642
Trevaskis B (2010) The central role of the VERNALIZATION1 gene in the vernalization response of cereals. Funct Plant Biol 37:479–487. https://doi.org/10.1071/FP10056
Vágújfalvi A, Galiba G, Cattivelli L, Dubcovsky J (2003) The cold-regulated transcriptional activator CBF3 is linked to the frost-tolerance locus Fr-A2 on wheat chromosome 5A. Mol Genet Genomics 269:60–67
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252. https://doi.org/10.1017/S0016672399004462
Williams E, Piepho HP, Whitaker D (2011) Augmented p-rep designs. Biom J 53:19–27. https://doi.org/10.1002/bimj.201000102
Worland A, Börner A, Korzun V, Li W, Petrovic S, Sayers E (1998) The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 100:385–394. https://doi.org/10.1023/A:1018327700985
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686. https://doi.org/10.1007/s00122-004-1796-4
Zadoks JC, Chang TT, Konzak CF (1974) A decimal code for the growth stages of cereals. Weed Res 14:415–421. https://doi.org/10.1111/j.1365-3180.1974.tb01084.x
Zhang X, Xiao Y, Zhang Y, Xia X, Dubcovsky J, He Z (2008) Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Sci 48:458–470. https://doi.org/10.2135/cropsci2007.06.0355
Zhang J, Wang Y, Wu S, Yang J, Liu H, Zhou Y (2012) A single nucleotide polymorphism at the Vrn-D1 promoter region in common wheat is associated with vernalization response. Theor Appl Genet 125:1697–1704. https://doi.org/10.1007/s00122-012-1946-z
Zhao Y, Gowda M, Würschum T, Longin CFH, Korzun V, Kollers S, Schachschneider R, Zeng J, Fernando R, Dubcovsky J (2013) Dissecting the genetic architecture of frost tolerance in central European winter wheat. J Exp Bot 64:4453–4460. https://doi.org/10.1093/jxb/ert259
Zhu J, Pearce S, Burke A, See DR, Skinner DZ, Dubcovsky J, Garland-Campbell K (2014) Copy number and haplotype variation at the VRN-A1 and central FR-A2 loci are associated with frost tolerance in hexaploid wheat. Theor Appl Genet 127:1183–1197. https://doi.org/10.1007/s00122-014-2290-2
Acknowledgements
The authors thank the USDA-ARS National Small Grains Collection for the timely delivery of seed used as controls for KASP analysis, Dr. Sarah Grogan for her help in compiling KASP primers, and Emily Hudson-Arns for helping to optimize KASP assays. The IWWIP is financially supported by Ministry of Food, Agriculture and Livestock of the Republic of Turkey and CRP WHEAT.
Author information
Authors and Affiliations
Contributions
CB designed the experiments, carried out collection of field phenotypes, conducted KASP marker analyses, performed all statistical analyses, and drafted the manuscript. VA conducted GBS library preparation and bioinformatics for SNP allele calling, coordinated KASP assay design, and provided editorial suggestions on the draft manuscript. SH helped to design and implement the experiments, assisted with data analysis, and provided editorial input on the draft manuscript. AM provided the germplasm used in these experiments and editorial input on the draft manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 337 kb)
Rights and permissions
About this article
Cite this article
Beil, C.T., Anderson, V.A., Morgounov, A. et al. Genomic selection for winter survival ability among a diverse collection of facultative and winter wheat genotypes. Mol Breeding 39, 29 (2019). https://doi.org/10.1007/s11032-018-0925-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-018-0925-8