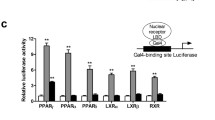
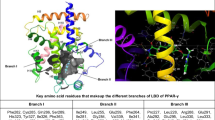
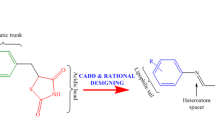
Abstract
PPARα and PPARγ are isoforms of the nuclear receptor superfamily which regulate glucose and lipid metabolism. Activation of PPARα and PPARγ receptors by exogenous ligands could transactivate the expression of PPARα and PPARγ-dependent genes, and thereby, metabolic pathways get triggered, which are helpful to ameliorate treatment for the type 2 diabetes mellitus, and related metabolic complications. Herein, by understanding the structural requirements for ligands to activate PPARα and PPARγ proteins, we developed a multilevel in silico-based virtual screening protocol to identify novel chemical scaffolds and further design and synthesize two distinct series of glitazone derivatives with advantages over the classical PPARα and PPARγ agonists. Moreover, the synthesized compounds were biologically evaluated for PPARα and PPARγ transactivation potency from nuclear extracts of 3T3-L1 cell. Furthermore, glucose uptake assay on L6 cells confirmed the potency of the synthesized compounds toward glucose regulation. Percentage lipid-lowering potency was also assessed through triglyceride estimate from 3T3-L1 cell extracts. Results suggested the ligand binding mode was in orthosteric fashion as similar to classical agonists. Thus molecular docking and molecular dynamics (MD) simulation experiments were executed to validate our hypothesis on mode of ligands binding and protein complex stability. Altogether, the present study developed a newer protocol for virtual screening and enables to design of novel glitazones for activation of PPARα and PPARγ-mediated pathways. Accordingly, present approach will offer benefit as a therapeutic strategy against type 2 diabetes mellitus and associated metabolic complications.
Graphical abstract

















Similar content being viewed by others
References
Yoon K, Lee J, Kim J et al (2006) Epidemic obesity and type 2 diabetes in Asia. Lancet 368(9548):1681–1688. https://doi.org/10.1016/S0140-6736(06)69703-1
Schulman IG (2010) Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev 62(13):1307–1315. https://doi.org/10.1016/j.addr.2010.07.002
Swanson HI, Wada T, Xie W et al (2013) Role of nuclear receptors in lipid dysfunction and obesity-related diseases. Drug Metab Dispos 41(1):1–11. https://doi.org/10.1124/dmd.112.048694
Dixon ED, Nardo AD, Claudel T, Trauner M (2021) The role of lipid sensing nuclear receptors (PPARs and LXR) and metabolic lipases in obesity. Diabetes NAFLD Genes (Basel) 12(5):645. https://doi.org/10.3390/genes12050645
Zhang C, Zhang B, Zhang X et al (2020) Targeting orphan nuclear receptors NR4As for energy homeostasis and diabetes. Front Pharmacol 11:1–11. https://doi.org/10.3389/fphar.2020.587457
Abdul M, Khan B, Hashim MJ et al (2020) Epidemiology of type 2 diabetes—global burden of disease and forecasted trends. J Epidemiol Glob Health 10(1):107–111. https://doi.org/10.2991/jegh.k.191028.001
Galicia-Garcia U, Benito-Vicente A, Jebari S et al (2020) Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci 21(17):1–34. https://doi.org/10.3390/ijms21176275
Rangwala SM, Lazar MA (2004) Peroxisome proliferator-activated receptor g in diabetes and metabolism. Trends Pharmacol Sci 25(6):331–336. https://doi.org/10.1016/j.tips.2004.03.012
O’Brien T, Nguyen TT, Zimmerman BR (1998) Hyperlipidemia and diabetes mellitus. Mayo Clin Proc 73(10):969–976. https://doi.org/10.4065/73.10.969
Fiévet C, Fruchart J-C, Staels B (2006) PPAR α and PPAR γ dual agonists for the treatment of type 2 diabetes and the metabolic syndrome. Curr Opin Pharmacol 6(6):606–614. https://doi.org/10.1016/j.coph.2006.06.009
Bajaj M, Suraamornkul S, Hardies LJ, Glass L (2007) Effects of peroxisome proliferator-activated receptor (PPAR) - α and PPAR- γ agonists on glucose and lipid metabolism in patients with type 2 diabetes mellitus. Diabetologia 50:1723–1731. https://doi.org/10.1007/s00125-007-0698-9
Kaur P, Rafiq Z, Bhat S et al (2020) Synthesis and evaluation of new 1, 2, 4-oxadiazole based trans—acrylic acid derivatives as potential PPAR-alpha/gamma dual agonist. Bioorg Chem 100:103867. https://doi.org/10.1016/j.bioorg.2020.103867
Yasmin S, Jayaprakash V (2017) Thiazolidinediones and PPAR orchestra as antidiabetic agents: from past to present. Eur J Med Chem 126:879–893. https://doi.org/10.1016/j.ejmech.2016.12.020
Darwish KM, Salama I, Mostafa S et al (2018) Synthesis, biological evaluation, and molecular docking investigation of benzhydrol- and indole-based dual PPAR-γ/FFAR1 agonists. Bioorganic Med Chem Lett 28(9):1595–1602. https://doi.org/10.1016/j.bmcl.2018.03.051
Lillich FF, Imig JD, Proschak E (2021) Multi-target approaches in metabolic syndrome. Front Pharmacol 11:1–18. https://doi.org/10.3389/fphar.2020.554961
González-álvarez H, Bravo-Jiménez A, Martínez-Arellanes M et al (2021) In silico-based design and in vivo evaluation of an anthranilic acid derivative as a multitarget drug in a diet-induced metabolic syndrome model. Pharmaceuticals 14(9):1–18. https://doi.org/10.3390/ph14090914
Feng X, Ding T, Liu Y et al (2021) In-silico Identification of peroxisome proliferator- activated receptor (PPAR)α/γ agonists from Ligand Expo Components database. J Biomol Struct Dyn 39(5):1853–1864. https://doi.org/10.1080/07391102.2020.1745279
Liu Y, Feng X, Jia W et al (2020) Virtual identification of novel PPARα/γ dual agonists by 3D-QSAR, molecule docking and molecular dynamics studies. J Biomol Struct Dyn 38(9):2672–2685. https://doi.org/10.1080/07391102.2019.1656110
Nath V, Agrawal R, Kumar V (2020) Structure based docking and molecular dynamics studies: peroxisome proliferator-activated receptors –α/γ dual agonists for treatment of metabolic disorders. J Biomol Struct Dyn 38(2):511–523. https://doi.org/10.1080/07391102.2019.1581089
Monsalve FA, Pyarasani RD, Delgado-Lopez F, Moore-Carrasco R (2013) Peroxisome proliferator-activated receptor targets for the treatment of metabolic diseases. Mediators Inflamm 549627:1–18. https://doi.org/10.1155/2013/549627
Corrales P, Vidal-Puig A, Medina-Gómez G (2018) PPARS and metabolic disorders associated with challenged adipose tissue plasticity. Int J Mol Sci 19(7):1–16. https://doi.org/10.3390/ijms19072124
Blaschke F, Takata Y, Caglayan E et al (2006) Obesity, peroxisome proliferator-activated receptor, and atherosclerosis in type 2 diabetes. Arterioscler Thromb Vasc Biol 26:28–40. https://doi.org/10.1161/01.ATV.0000191663.12164.77
Tsukidate T, Li Q, Hang HC (2020) Nuclear receptor chemical reporter enables domain-specific analysis of ligands in mammalian cells. ACS Chem Biol 15(9):2324–2330. https://doi.org/10.1021/acschembio.0c00432
Brust R, Lin H, Fuhrmann J et al (2017) Modification of the orthosteric PPARγ covalent antagonist scaffold yields an improved dual-site allosteric inhibitor. ACS Chem Biol 12(4):969–978. https://doi.org/10.1021/acschembio.6b01015
Chandra V, Huang P, Hamuro Y et al (2008) Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature 456:350–356. https://doi.org/10.1038/nature07413
Kumar S, Suleski MP, Markov GJ et al (2009) Positional conservation and amino acids shape the correct diagnosis and population frequencies of benign and damaging personal amino acid mutations. Genome Res 19:1562–1569. https://doi.org/10.1101/gr.091991.109
Biro JC (2006) Amino acid size, charge, hydropathy indices and matrices for protein structure analysis. Theor Biol Med Model 3:1–12. https://doi.org/10.1186/1742-4682-3-15
Sarwar N, Gao P, Kondapally Seshasai SR et al (2010) Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet 375(9733):2215–2222. https://doi.org/10.1016/S0140-6736(10)60484-9
Stadler LKJ, Farooqi IS (2017) A new drug target for type 2 diabetes. Cell 170(1):12–14. https://doi.org/10.1016/j.cell.2017.06.024
Karalliedde J, Buckingham RE (2007) Thiazolidinediones and their fluid-related adverse effects: facts, fiction and putative management strategies. Drug Saf 30:741–753. https://doi.org/10.2165/00002018-200730090-00002
Hussein Z, Wentworth JM, Nankervis AJ et al (2004) Effectiveness and side effects of thiazolidinediones for type 2 diabetes: real-life experience from a tertiary hospital. Med J Aust 181(10):536–539. https://doi.org/10.5694/j.1326-5377.2004.tb06441.x
Hedrington MS, Davis SN (2018) Peroxisome proliferator-activated receptor alpha-mediated drug toxicity in the liver. Expert Opin Drug Metab Toxicol 14(7):671–677. https://doi.org/10.1080/17425255.2018.1483337
Khuchua Z, Glukhov AI, Strauss AW, Javadov S (2018) Elucidating the beneficial role of PPAR agonists in cardiac diseases. Int J Mol Sci 19(11):1–17. https://doi.org/10.3390/ijms19113464
Shi R, Zhao L, Qi Y (2018) The effect of fenofibrate on early retinal nerve fiber layer loss in type 2 diabetic patients: a case-control study. BMC Ophthalmol 18(1):100. https://doi.org/10.1186/s12886-018-0769-3
Rubenstrunk A, Hanf R, Hum DW et al (2007) Safety issues and prospects for future generations of PPAR modulators. Biochim Biophys Acta 1771(8):1065–1081. https://doi.org/10.1016/j.bbalip.2007.02.003
Balakumar P, Mahadevan N, Sambathkumar R (2019) A contemporary overview of PPARα/γ dual agonists for the management of diabetic dyslipidemia. Curr Mol Pharmacol 12(3):195–201. https://doi.org/10.2174/1874467212666190111165015
Li J, Kennedy LJ, Shi Y et al (2010) Discovery of an oxybenzylglycine based peroxisome proliferator activated receptor α selective agonist 2-((3-((2-(4-chlorophenyl)-5-methyloxazol-4- yl)methoxy)benzyl)(methoxycarbonyl)amino)acetic acid (BMS-687453). J Med Chem 53(7):2854–2864. https://doi.org/10.1021/jm9016812
Nolte RT, Wisely GB, Westin S et al (1998) Ligand binding and co-activator assembly of the peroxisome proliferator- activated receptor-γ. Nature 395:137–143. https://doi.org/10.1038/25931
Chen JH, Linstead E, Swamidass SJ et al (2007) ChemDB update—full-text search and virtual chemical space. Bioinformatics 23(17):2348–2351. https://doi.org/10.1093/bioinformatics/btm341
Fu J, Si P, Zheng M et al (2012) Discovery of new non-steroidal FXR ligands via a virtual screening workflow based on Phase shape and induced fit docking. Bioorg Med Chem Lett 22(22):6848–6853. https://doi.org/10.1016/j.bmcl.2012.09.045
Kowalska M, Fijałkowski Ł, Nowaczyk A (2018) The biological activity assessment of potential drugs acting on cardiovascular system using Lipinski and Veber Rules. J Educ Heal Sport 8(12):184–191. https://doi.org/10.5281/zenodo.2066519
Chen X, Li H, Tian L et al (2020) Analysis of the physicochemical properties of acaricides based on Lipinski’s rule of five. J Comput Biol 27(9):1–10. https://doi.org/10.1089/cmb.2019.0323
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690. https://doi.org/10.1002/jcc.21367
Rao SN, Head MS, Kulkarni A et al (2007) Validation studies of the site-directed docking program LibDock. J Chem Inf Model 47(6):2159–2171. https://doi.org/10.1021/ci6004299
Wu G, Robertson DH, Brooks CL III, Vieth M (2003) Detailed analysis of grid-based molecular docking: a case study of CDOCKER - A CHARMm based MD docking program. J Comput Chem 24(13):1549–1562. https://doi.org/10.1002/jcc.10306
Markt P, Petersen RK, Flindt EN et al (2008) Discovery of novel PPAR ligands by a virtual screening approach based on pharmacophore modeling, 3D shape, and electrostatic similarity screening. J Med Chem 51(20):6303–6317. https://doi.org/10.1021/jm800128k
Toledo Warshaviak D, Golan G, Borrelli KW et al (2014) Structure-based virtual screening approach for discovery of covalently bound ligands. J Chem Inf Model 54(7):1941–1950. https://doi.org/10.1021/ci500175r
Xu X, Huang M, Zou X (2018) Docking-based inverse virtual screening: methods, applications, and challenges. Biophys Reports 4:1–16. https://doi.org/10.1007/s41048-017-0045-8
Melo-Filho C, Braga R, Andrade C (2014) 3D-QSAR approaches in drug design: perspectives to generate reliable CoMFA models. Curr Comput Aided-Drug Des 10(2):148–159. https://doi.org/10.2174/1573409910666140410111043
Cheng Y, Zhou M, Tung CH et al (2010) Studies on two types of PTP1B inhibitors for the treatment of type 2 diabetes: Hologram QSAR for OBA and BBB analogues. Bioorganic Med Chem Lett 20(11):3329–3337. https://doi.org/10.1016/j.bmcl.2010.04.033
Freitas RF, Oprea TI, Montanari C, a. (2008) 2D QSAR and similarity studies on cruzain inhibitors aimed at improving selectivity over cathepsin L. Bioorganic Med Chem 16(2):838–853. https://doi.org/10.1016/j.bmc.2007.10.048
Daura X, Gademann K, Jaun B et al (1999) Peptide folding: when simulation meets experiment. Angew Chem Int Ed 38:236–240. https://doi.org/10.1002/(SICI)1521-3773(19990115)38:1/2%3c236::AID-ANIE236%3e3.0.CO;2-M
Lyubartsev AP, Laaksonen A (2000) M. DynaMix—a scalable portable parallel MD simulation package for arbitrary molecular mixtures. Comput Phys Commun 128(3):565–589. https://doi.org/10.1016/S0010-4655(99)00529-9
Tang YW, Huang Z, Wang X, Zeng XC (2006) Molecular dynamics simulations of thermal conductivity of silicon nanotubes. J Comput Theor Nanosci 3(5):824–829. https://doi.org/10.1166/jctn.2006.023
Yamamoto N, Kawasaki K, Sato T et al (2008) A nonradioisotope, enzymatic microplate assay for in vivo evaluation of 2-deoxyglucose uptake in muscle tissue. Anal Biochem 375(2):397–399. https://doi.org/10.1016/j.ab.2008.01.013
Yamamoto N, Sato T, Kawasaki K et al (2006) A nonradioisotope, enzymatic assay for 2-deoxyglucose uptake in L6 skeletal muscle cells cultured in a 96-well microplate. Anal Biochem 351(1):139–145. https://doi.org/10.1016/j.ab.2005.12.011
Das MS, Devi G (2015) In vitro cytotoxicity and glucose uptake activity of fruits of terminalia bellirica in Vero, L-6 and 3T3 cell lines. J Appl Pharm Sci 5(12):92–95. https://doi.org/10.7324/JAPS.2015.501215
Watanabe K, Matsumoto A, Tsuda H, Iwamoto S (2021) N4BP2L1 interacts with dynactin and contributes to GLUT4 trafficking and glucose uptake in adipocytes. J Diabetes Investig 12:1958–1966. https://doi.org/10.1111/jdi.13623
Chen D, Wang H, Chen J et al (2018) MicroRNA-129-5p regulates glycolysis and cell proliferation by targeting the glucose transporter SLC2A3 in gastric cancer cells. Front Pharmacol 9:1–10. https://doi.org/10.3389/fphar.2018.00502
Barik SK, Russell WR, Moar KM et al (2020) The anthocyanins in black currants regulate postprandial hyperglycaemia primarily by inhibiting α-glucosidase while other phenolics modulate salivary α-amylase, glucose uptake and sugar transporters. J Nutr Biochem 78:108325. https://doi.org/10.1016/j.jnutbio.2019.108325
Müller MS, Fouyssac M, Taylor CW (2018) Effective Glucose uptake by human astrocytes requires its sequestration in the endoplasmic reticulum by glucose-6-phosphatase-β. Curr Biol 28(21):3481–3486. https://doi.org/10.1016/j.cub.2018.08.060
Karunakaran RS, Lokanatha O, Swamy GM, Miranda V (2021) Anti-obesity and lipid lowering activity of bauhiniastatin-1 is mediated through PPAR- γ/AMPK expressions in diet-induced obese rat model. Front Pharmacol 12:1–13. https://doi.org/10.3389/fphar.2021.704074
Iwamoto K, Kamo S, Takada Y et al (2018) Soyasapogenols reduce cellular triglyceride levels in 3T3-L1 mouse adipocyte cells by accelerating triglyceride lipolysis. Biochem Biophys Reports 16:44–49. https://doi.org/10.1016/j.bbrep.2018.09.006
Feng C, Li D, Chen M et al (2019) Citreoviridin induces myocardial apoptosis through PPAR- γ -mTORC2- mediated autophagic pathway and the protective effect of thiamine and selenium. Chem Biol Interact 311:108795. https://doi.org/10.1016/j.cbi.2019.108795
Jin M, Lai Y, Zhang W (2020) Effects of peptidoglycan on the development of steatohepatitis. BBA 1865:158595. https://doi.org/10.1016/j.bbalip.2019.158595
Gersch M, Gladkova C, Schubert AF et al (2017) Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat Struct Mol Biol 24:920–930. https://doi.org/10.1038/nsmb.3475
Van Der Spoel D, Lindahl E, Hess B et al (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Hess B, Kutzner C, Van Der Spoel D, Lindahl E (2008) GRGMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447. https://doi.org/10.1021/ct700301q
Abraham MJ, Murtola T, Schulz R et al (2015) Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Larsson P, Cuendet MA, Hess B, Lindahl E (2010) Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water. J Chem Theory Comput 6(2):459–466. https://doi.org/10.1021/ct900549r
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC / E water models at 298 K. J Phys Chem A 105(43):9954–9960. https://doi.org/10.1021/jp003020w
Wang T, Brudvig GW, Batista VS (2010) Study of proton coupled electron transfer in a biomimetic dimanganese water oxidation catalyst with terminal water ligands. J Chem Theory Comput 6(8):2395–2401. https://doi.org/10.1021/ct1002658
Mandal SP, Garg A, Prabitha P et al (2018) Novel glitazones as PPARγ agonists: molecular design, synthesis, glucose uptake activity and 3D QSAR studies. Chem Cent J 12:141. https://doi.org/10.1186/s13065-018-0508-0
Leach AR, Gillet VJ, Lewis RA, Taylor R (2010) Three-dimensional pharmacophore methods in drug discovery. J Med Chem 53(2):539–558. https://doi.org/10.1021/jm900817u
Yang S (2010) Pharmacophore modeling and applications in drug discovery : challenges and recent advances. Drug Discov Today 15:444–450. https://doi.org/10.1016/j.drudis.2010.03.013
Heravi MM, Ghods A, Bakhtiari K, Derikvand F (2010) Zn[(L)proline]2: an efficient catalyst for the synthesis of biologically active pyrano[2,3-d]pyrimidine derivatives. Synth Commun 40(13):1927–1931. https://doi.org/10.1080/00397910903174390
Singh I, Rani R, Luxami V, Paul K (2019) Synthesis of 5-(4-(1H-phenanthro[9,10-d]imidazol-2-yl)benzylidene)thiazolidine-2,4-dione as promising DNA and serum albumin-binding agents and evaluation of antitumor activity. Eur J Med Chem 166:267–280. https://doi.org/10.1016/j.ejmech.2019.01.053
Stepanek O, Hin N, Thomas AG et al (2019) Neutral sphingomyelinase 2 inhibitors based on the 4-(1H-imidazol-2-yl)-2,6-dialkoxyphenol scaffold. Eur J Med Chem 170:276–289. https://doi.org/10.1016/j.ejmech.2019.03.015
Frédérick R, Robert S, Charlier C et al (2007) Mechanism-based thrombin inhibitors: design, synthesis, and molecular docking of a new selective 2-oxo-2H-1-benzopyran derivative. J Med Chem 50(15):3645–3650. https://doi.org/10.1021/jm061368v
Mandal S, Br PK, Alam T et al (2022) Novel imidazole phenoxyacetic acids as inhibitors of USP30 for neuroprotection implication via the ubiquitin-Rho-110 fluorometric assay: design, synthesis, and in silico and biochemical assays. ACS Chem Neurosci 13(9):1433–1445. https://doi.org/10.1021/acschemneuro.2c00076
Lin Y, Huang J, Wu C et al (2002) Design, synthesis, and evaluation of postulated transient intermediate and substrate analogues as inhibitors of 4-hydroxyphenylpyruvate dioxygenase. Bioorg Med Chem Lett 12(13):1709–1713. https://doi.org/10.1016/S0960-894X(02)00291-3
Trapero A, Pacitto A, Singh V et al (2018) A fragment-based approach to targeting inosine-5 ´—monophosphate dehydrogenase ( IMPDH ) from mycobacterium tuberculosis. J Med Chem 61(7):2806–2822. https://doi.org/10.1021/acs.jmedchem.7b01622
Wang Y, Wu G, Xu X et al (2021) Palladium-Catalyzed β-C(sp 3)-H Arylation of aliphatic ketones enabled by a transient directing group. J Org Chem 86(10):7296–7303. https://doi.org/10.1021/acs.joc.1c00646
He G, Song Q, Wang J et al (2020) Design, synthesis and biological evaluation of N -hydroxy- aminobenzyloxyarylamide analogues as novel selective κ opioid receptor antagonists. Bioorg Med Chem Lett 30(13):127236. https://doi.org/10.1016/j.bmcl.2020.127236
Lin H, Pan X, Barsamian AL et al (2019) Native directed site-selective δ - C(sp 3)-H and δ-C(sp 2)-H Arylation of primary amines. ACS Catal 9(6):4887–4891. https://doi.org/10.1021/acscatal.8b04927
Prem A, Mandal S, Prabitha P, Faizan S (2022) Rational design, molecular docking, dynamic simulation, synthesis, PPAR- γ competitive binding and transcription analysis of novel glitazones. J Mol Struct 1265:133354. https://doi.org/10.1016/j.molstruc.2022.133354
Musser JH, Kubrak DM, Chang J, et al (1987) Leukotriene D4 antagonists and 5-lipoxygenase inhibitors. Synthesis of benzoheterocyclic [(methoxyphenyl)amino] oxoalkanoic acid esters. J Med Chem 30(2):400–405. https://doi.org/10.1021/jm00385a024
Lundin A, Hasenson M, Persson J, Pousette A (1986) Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol 133:27–42. https://doi.org/10.1016/0076-6879(86)33053-2
Gonzalez RJ, Tarloff JB (2001) Evaluation of hepatic subcellular fractions for Alamar blue and MTT reductase activity. Toxicol Vitr 15(3):257–259. https://doi.org/10.1016/S0887-2333(01)00014-5
Denizot F, Lang R (1986) Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods 89(2):271–277. https://doi.org/10.1016/0022-1759(86)90368-6
Nisha CM, Kumar A, Nair P et al (2016) Molecular docking and in silico ADMET study reveals acylguanidine 7a as a potential inhibitor of β -secretase. Adv Bioinformatics 2016:1–6. https://doi.org/10.1155/2016/9258578
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:1–13. https://doi.org/10.1038/srep42717
Dong J, Wang NN, Yao ZJ et al (2018) Admetlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminform 10:1–11. https://doi.org/10.1186/s13321-018-0283-x
Acknowledgements
Subhankar Mandal is thankful to DST INSPIRE, Department of Science and Technology (DST), New Delhi, India, for doctoral grant and financial support in the form of Research Fellowship (DST/INSPIRE Fellowship/2016/IF160630). Authors are thankful to the management, JSS academy of higher education and research, Mysore, for providing necessary support.
Funding
Funding was provided by DST Inspire India (Grant Number IF160630).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no competing interest in any form.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mandal, S., Faizan, S., Raghavendra, N.M. et al. Molecular dynamics articulated multilevel virtual screening protocol to discover novel dual PPAR α/γ agonists for anti-diabetic and metabolic applications. Mol Divers 27, 2605–2631 (2023). https://doi.org/10.1007/s11030-022-10571-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-022-10571-w